Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance
- PMID: 19478082
- PMCID: PMC2740434
- DOI: 10.1074/jbc.M109.018010
Multiple conformers in active site of human dihydrofolate reductase F31R/Q35E double mutant suggest structural basis for methotrexate resistance
Abstract
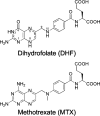
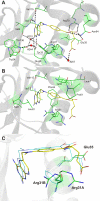
Methotrexate is a slow, tight-binding, competitive inhibitor of human dihydrofolate reductase (hDHFR), an enzyme that provides key metabolites for nucleotide biosynthesis. In an effort to better characterize ligand binding in drug resistance, we have previously engineered hDHFR variant F31R/Q35E. This variant displays a >650-fold decrease in methotrexate affinity, while maintaining catalytic activity comparable to the native enzyme. To elucidate the molecular basis of decreased methotrexate affinity in the doubly substituted variant, we determined kinetic and inhibitory parameters for the simple variants F31R and Q35E. This demonstrated that the important decrease of methotrexate affinity in variant F31R/Q35E is a result of synergistic effects of the combined substitutions. To better understand the structural cause of this synergy, we obtained the crystal structure of hDHFR variant F31R/Q35E complexed with methotrexate at 1.7-A resolution. The mutated residue Arg-31 was observed in multiple conformers. In addition, seven native active-site residues were observed in more than one conformation, which is not characteristic of the wild-type enzyme. This suggests that increased residue disorder underlies the observed methotrexate resistance. We observe a considerable loss of van der Waals and polar contacts with the p-aminobenzoic acid and glutamate moieties. The multiple conformers of Arg-31 further suggest that the amino acid substitutions may decrease the isomerization step required for tight binding of methotrexate. Molecular docking with folate corroborates this hypothesis.
Figures

References
-
- Slamon D. J., Romond E. H., Perez E. A. (2006) Clin. Adv. Hematol. Oncol. 4, Suppl. 1, 4–9 - PubMed
-
- Daw N. C., Billups C. A., Rodriguez-Galindo C., McCarville M. B., Rao B. N., Cain A. M., Jenkins J. J., Neel M. D., Meyer W. H. (2006) Cancer 106, 403–412 - PubMed
-
- Strojan P., Soba E., Budihna M., Auersperg M. (2005) J. Surg. Oncol. 92, 278–283 - PubMed
-
- Flintoff W. F., Sadlish H., Gorlick R., Yang R., Williams F. M. (2004) Biochim. Biophys. Acta 1690, 110–117 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

