Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa
- PMID: 19478873
- PMCID: PMC2680969
- DOI: 10.1371/journal.ppat.1000455
Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa
Abstract
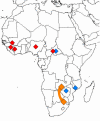
Lujo virus (LUJV), a new member of the family Arenaviridae and the first hemorrhagic fever-associated arenavirus from the Old World discovered in three decades, was isolated in South Africa during an outbreak of human disease characterized by nosocomial transmission and an unprecedented high case fatality rate of 80% (4/5 cases). Unbiased pyrosequencing of RNA extracts from serum and tissues of outbreak victims enabled identification and detailed phylogenetic characterization within 72 hours of sample receipt. Full genome analyses of LUJV showed it to be unique and branching off the ancestral node of the Old World arenaviruses. The virus G1 glycoprotein sequence was highly diverse and almost equidistant from that of other Old World and New World arenaviruses, consistent with a potential distinctive receptor tropism. LUJV is a novel, genetically distinct, highly pathogenic arenavirus.
Conflict of interest statement
SKH and ME are employees of 454 Life Sciences, Inc., a Roche Company.
Figures


References
-
- Bowen MD, Peters CJ, Nichol ST. Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol. 1997;8:301–316. - PubMed
-
- Moncayo AC, Hice CL, Watts DM, Travassos de Rosa AP, Guzman H, et al. Allpahuayo virus: a newly recognized arenavirus (arenaviridae) from arboreal rice rats (oecomys bicolor and oecomys paricola) in northeastern peru. Virology. 2001;284:277–286. - PubMed
-
- Armstrong C, Lillie RD. Experimental lymphocytic choriomeningitis of monkeys and mice produced by a virus encountered in studies of the 1933 St. Louis encephalitis epidemic. Public Health Rep. 1934;49:1019–1027.
-
- Salvato MS, Shimomaye EM. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology. 1989;173:1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

