Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication
- PMID: 19478885
- PMCID: PMC2682650
- DOI: 10.1371/journal.ppat.1000462
Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication
Abstract
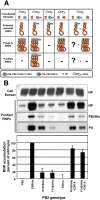
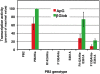
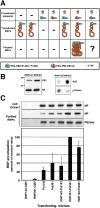
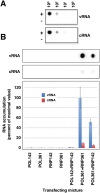
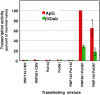
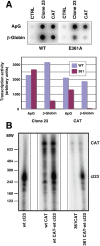
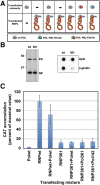
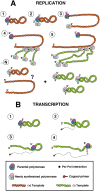
The influenza A viruses genome comprises eight single-stranded RNA segments of negative polarity. Each one is included in a ribonucleoprotein particle (vRNP) containing the polymerase complex and a number of nucleoprotein (NP) monomers. Viral RNA replication proceeds by formation of a complementary RNP of positive polarity (cRNP) that serves as intermediate to generate many progeny vRNPs. Transcription initiation takes place by a cap-snatching mechanism whereby the polymerase steals a cellular capped oligonucleotide and uses it as primer to copy the vRNP template. Transcription termination occurs prematurely at the polyadenylation signal, which the polymerase copies repeatedly to generate a 3'-terminal polyA. Here we studied the mechanisms of the viral RNA replication and transcription. We used efficient systems for recombinant RNP transcription/replication in vivo and well-defined polymerase mutants deficient in either RNA replication or transcription to address the roles of the polymerase complex present in the template RNP and newly synthesised polymerase complexes during replication and transcription. The results of trans-complementation experiments showed that soluble polymerase complexes can synthesise progeny RNA in trans and become incorporated into progeny vRNPs, but only transcription in cis could be detected. These results are compatible with a new model for virus RNA replication, whereby a template RNP would be replicated in trans by a soluble polymerase complex and a polymerase complex distinct from the replicative enzyme would direct the encapsidation of progeny vRNA. In contrast, transcription of the vRNP would occur in cis and the resident polymerase complex would be responsible for mRNA synthesis and polyadenylation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Ultrastructure of influenza virus ribonucleoprotein complexes during viral RNA synthesis.Commun Biol. 2021 Jul 9;4(1):858. doi: 10.1038/s42003-021-02388-4. Commun Biol. 2021. PMID: 34244608 Free PMC article.
-
The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication.Acta Virol. 2013;57(2):113-22. doi: 10.4149/av_2013_02_113. Acta Virol. 2013. PMID: 23600869 Review.
-
Structural and functional characterization of an influenza virus RNA polymerase-genomic RNA complex.J Virol. 2010 Oct;84(20):10477-87. doi: 10.1128/JVI.01115-10. Epub 2010 Aug 11. J Virol. 2010. PMID: 20702645 Free PMC article.
-
Transcription and replication of the influenza a virus genome.Acta Virol. 2000 Oct;44(5):273-82. Acta Virol. 2000. PMID: 11252672 Review.
-
Comparison of two reconstituted systems for in vitro transcription and replication of influenza virus.J Biochem. 1992 Apr;111(4):496-9. doi: 10.1093/oxfordjournals.jbchem.a123786. J Biochem. 1992. PMID: 1618740
Cited by
-
Diversity and Complexity of Internally Deleted Viral Genomes in Influenza A Virus Subpopulations with Enhanced Interferon-Inducing Phenotypes.Viruses. 2023 Oct 17;15(10):2107. doi: 10.3390/v15102107. Viruses. 2023. PMID: 37896883 Free PMC article.
-
Influenza a virus host shutoff disables antiviral stress-induced translation arrest.PLoS Pathog. 2014 Jul 10;10(7):e1004217. doi: 10.1371/journal.ppat.1004217. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25010204 Free PMC article.
-
At the centre: influenza A virus ribonucleoproteins.Nat Rev Microbiol. 2015 Jan;13(1):28-41. doi: 10.1038/nrmicro3367. Epub 2014 Nov 24. Nat Rev Microbiol. 2015. PMID: 25417656 Free PMC article. Review.
-
Ultrastructure of influenza virus ribonucleoprotein complexes during viral RNA synthesis.Commun Biol. 2021 Jul 9;4(1):858. doi: 10.1038/s42003-021-02388-4. Commun Biol. 2021. PMID: 34244608 Free PMC article.
-
Biological activities of 'noninfectious' influenza A virus particles.Future Virol. 2014 Jan;9(1):41-51. doi: 10.2217/fvl.13.118. Future Virol. 2014. PMID: 25067941 Free PMC article.
References
-
- Wright PF, Neumann G, Kawaoka Y. Orthomyxoviridae. In: Knipe DM, Howley PM, editors. Fields Virology 5th edition. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1691–1740.
-
- Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005;3:591–600. - PubMed
-
- Palese P, Shaw M. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology 5th edition. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1647–1689.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

