Transcriptome-wide prediction of miRNA targets in human and mouse using FASTH
- PMID: 19478946
- PMCID: PMC2684643
- DOI: 10.1371/journal.pone.0005745
Transcriptome-wide prediction of miRNA targets in human and mouse using FASTH
Erratum in
- PLoS One. 2009;4(7). doi: 10.1371/annotation/e0842765-3cae-4737-8b5b-96aeb12d7fb5
Abstract
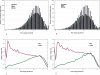
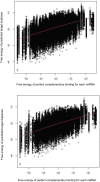
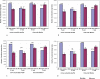
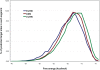
Transcriptional regulation by microRNAs (miRNAs) involves complementary base-pairing at target sites on mRNAs, yielding complex secondary structures. Here we introduce an efficient computational approach and software (FASTH) for genome-scale prediction of miRNA target sites based on minimizing the free energy of duplex structure. We apply our approach to identify miRNA target sites in the human and mouse transcriptomes. Our results show that short sequence motifs in the 5' end of miRNAs frequently match mRNAs perfectly, not only at validated target sites but additionally at many other, energetically favourable sites. High-quality matching regions are abundant and occur at similar frequencies in all mRNA regions, not only the 3'UTR. About one-third of potential miRNA target sites are reassigned to different mRNA regions, or gained or lost altogether, among different transcript isoforms from the same gene. Many potential miRNA target sites predicted in human are not found in mouse, and vice-versa, but among those that do occur in orthologous human and mouse mRNAs most are situated in corresponding mRNA regions, i.e. these sites are themselves orthologous. Using a luciferase assay in HEK293 cells, we validate four of six predicted miRNA-mRNA interactions, with the mRNA level reduced by an average of 73%. We demonstrate that a thermodynamically based computational approach to prediction of miRNA binding sites on mRNAs can be scaled to analyse complete mammalian transcriptome datasets. These results confirm and extend the scope of miRNA-mediated species- and transcript-specific regulation in different cell types, tissues and developmental conditions.
Conflict of interest statement
Figures



Similar articles
-
Computational analysis of microRNA targets in Caenorhabditis elegans.Gene. 2006 Jan 3;365:2-10. doi: 10.1016/j.gene.2005.09.035. Epub 2005 Dec 13. Gene. 2006. PMID: 16356665
-
The human transcriptome is enriched for miRNA-binding sites located in cooperativity-permitting distance.RNA Biol. 2013 Jul;10(7):1125-35. doi: 10.4161/rna.24955. Epub 2013 May 9. RNA Biol. 2013. PMID: 23696004 Free PMC article.
-
The Binding Sites of miR-619-5p in the mRNAs of Human and Orthologous Genes.BMC Genomics. 2017 Jun 1;18(1):428. doi: 10.1186/s12864-017-3811-6. BMC Genomics. 2017. PMID: 28569192 Free PMC article.
-
miRNA Targeting: Growing beyond the Seed.Trends Genet. 2019 Mar;35(3):215-222. doi: 10.1016/j.tig.2018.12.005. Epub 2019 Jan 9. Trends Genet. 2019. PMID: 30638669 Free PMC article. Review.
-
MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions.Mol Cells. 2016 May 31;39(5):375-81. doi: 10.14348/molcells.2016.0013. Epub 2016 Apr 27. Mol Cells. 2016. PMID: 27117456 Free PMC article. Review.
Cited by
-
Re-thinking miRNA-mRNA interactions: intertwining issues confound target discovery.Bioessays. 2015 Apr;37(4):379-88. doi: 10.1002/bies.201400191. Epub 2015 Feb 12. Bioessays. 2015. PMID: 25683051 Free PMC article.
-
Gene bi-targeting by viral and human miRNAs.BMC Bioinformatics. 2010 May 13;11:249. doi: 10.1186/1471-2105-11-249. BMC Bioinformatics. 2010. PMID: 20465802 Free PMC article.
-
Comprehensive Analysis of Human microRNA-mRNA Interactome.Front Genet. 2019 Oct 8;10:933. doi: 10.3389/fgene.2019.00933. eCollection 2019. Front Genet. 2019. PMID: 31649721 Free PMC article.
-
Optimization of signal-to-noise ratio for efficient microarray probe design.Bioinformatics. 2016 Sep 1;32(17):i552-i558. doi: 10.1093/bioinformatics/btw451. Bioinformatics. 2016. PMID: 27587674 Free PMC article.
-
Temporal analysis of reciprocal miRNA-mRNA expression patterns predicts regulatory networks during differentiation in human skeletal muscle cells.Physiol Genomics. 2015 Mar;47(3):45-57. doi: 10.1152/physiolgenomics.00037.2014. Epub 2014 Dec 29. Physiol Genomics. 2015. PMID: 25547110 Free PMC article.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Lewis BP, Burge C, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNAs targets. Cell. 2005;120:15–20. - PubMed
-
- Manfield JH, Harf BD, Nissen R, Obenauer J, Srineel J, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

