Expanded alternative splice isoform profiling of the mouse Cav3.1/alpha1G T-type calcium channel
- PMID: 19480703
- PMCID: PMC2696442
- DOI: 10.1186/1471-2199-10-53
Expanded alternative splice isoform profiling of the mouse Cav3.1/alpha1G T-type calcium channel
Abstract
Background: Alternative splicing of low-voltage-activated T-type calcium channels contributes to the molecular and functional diversity mediating complex network oscillations in the normal brain. Transcript scanning of the human CACNA1G gene has revealed the presence of 11 regions within the coding sequence subjected to alternative splicing, some of which enhance T-type current. In mouse models of absence epilepsy, elevated T-type calcium currents without clear increases in channel expression are found in thalamic neurons that promote abnormal neuronal synchronization. To test whether enhanced T-type currents in these models reflect pathogenic alterations in channel splice isoforms, we determined the extent of alternative splicing of mouse Cacna1g transcripts and whether evidence of altered transcript splicing could be detected in mouse absence epilepsy models.
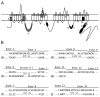
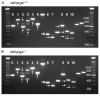
Results: Transcript scanning of the murine Cacna1g gene detected 12 regions encoding alternative splice isoforms of Cav3.1/alpha1G T-type calcium channels. Of the 12 splice sites, six displayed homology to the human CACNA1G splice sites, while six novel mouse-specific splicing events were identified, including one intron retention, three alternative acceptor sites, one alternative donor site, and one exon exclusion. In addition, two brain region-specific alternative splice patterns were observed in the cerebellum. Comparative analyses of brain regions from four monogenic absence epilepsy mouse models with altered thalamic T-type currents and wildtype controls failed to reveal differences in Cacna1g splicing patterns.
Conclusion: The determination of six novel alternative splice sites within the coding region of the mouse Cacna1g gene greatly expands the potential biophysical diversity of voltage-gated T-type channels in the mouse central nervous system. Although alternative splicing of Cav3.1/alpha1G channels does not explain the enhancement of T-type current identified in four mouse models of absence epilepsy, post-transcriptional modification of T-type channels through this mechanism may influence other developmental neurological phenotypes.
Figures




Similar articles
-
Genetic enhancement of thalamocortical network activity by elevating alpha 1g-mediated low-voltage-activated calcium current induces pure absence epilepsy.J Neurosci. 2009 Feb 11;29(6):1615-25. doi: 10.1523/JNEUROSCI.2081-08.2009. J Neurosci. 2009. PMID: 19211869 Free PMC article.
-
Structure and alternative splicing of the gene encoding alpha1G, a human brain T calcium channel alpha1 subunit.Neurosci Lett. 1999 Oct 29;274(3):143-6. doi: 10.1016/s0304-3940(99)00716-8. Neurosci Lett. 1999. PMID: 10548410
-
Functional impact of alternative splicing of human T-type Cav3.3 calcium channels.J Neurophysiol. 2004 Dec;92(6):3399-407. doi: 10.1152/jn.00498.2004. Epub 2004 Jul 14. J Neurophysiol. 2004. PMID: 15254077
-
Molecular characterization of T-type calcium channels.Cell Calcium. 2006 Aug;40(2):89-96. doi: 10.1016/j.ceca.2006.04.012. Epub 2006 Jun 8. Cell Calcium. 2006. PMID: 16759699 Review.
-
Characterization of the gating brake in the I-II loop of CaV3 T-type calcium channels.Channels (Austin). 2010 Nov-Dec;4(6):453-8. doi: 10.4161/chan.4.6.12889. Epub 2010 Nov 1. Channels (Austin). 2010. PMID: 21099341 Free PMC article. Review.
Cited by
-
Neuronal Cav3 channelopathies: recent progress and perspectives.Pflugers Arch. 2020 Jul;472(7):831-844. doi: 10.1007/s00424-020-02429-7. Epub 2020 Jul 7. Pflugers Arch. 2020. PMID: 32638069 Free PMC article. Review.
-
Calcium Channel CaVα₁ Splice Isoforms - Tissue Specificity and Drug Action.Curr Mol Pharmacol. 2015;8(1):22-31. doi: 10.2174/1874467208666150507103215. Curr Mol Pharmacol. 2015. PMID: 25966698 Free PMC article. Review.
-
Inhibition of T-Type calcium channels in mEC layer II stellate neurons reduces neuronal hyperexcitability associated with epilepsy.Epilepsy Res. 2019 Aug;154:132-138. doi: 10.1016/j.eplepsyres.2019.05.006. Epub 2019 May 18. Epilepsy Res. 2019. PMID: 31132598 Free PMC article.
-
Bordetella Dermonecrotic Toxin Is a Neurotropic Virulence Factor That Uses CaV3.1 as the Cell Surface Receptor.mBio. 2020 Mar 24;11(2):e03146-19. doi: 10.1128/mBio.03146-19. mBio. 2020. PMID: 32209694 Free PMC article.
-
Confirmation of an epilepsy modifier locus on mouse chromosome 11 and candidate gene analysis by RNA-Seq.Genes Brain Behav. 2012 Jun;11(4):452-60. doi: 10.1111/j.1601-183X.2012.00790.x. Epub 2012 Apr 27. Genes Brain Behav. 2012. PMID: 22471526 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

