Cytokine-dependent and-independent gene expression changes and cell cycle block revealed in Trypanosoma cruzi-infected host cells by comparative mRNA profiling
- PMID: 19480704
- PMCID: PMC2709661
- DOI: 10.1186/1471-2164-10-252
Cytokine-dependent and-independent gene expression changes and cell cycle block revealed in Trypanosoma cruzi-infected host cells by comparative mRNA profiling
Abstract
Background: The requirements for growth and survival of the intracellular pathogen Trypanosoma cruzi within mammalian host cells are poorly understood. Transcriptional profiling of the host cell response to infection serves as a rapid read-out for perturbation of host physiology that, in part, reflects adaptation to the infective process. Using Affymetrix oligonucleotide array analysis we identified common and disparate host cell responses triggered by T. cruzi infection of phenotypically diverse human cell types.
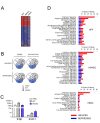
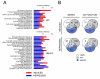
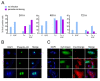
Results: We report significant changes in transcript abundance in T. cruzi-infected fibroblasts, endothelial cells and smooth muscle cells (2852, 2155 and 531 genes respectively; fold-change > or = 2, p-value < 0.01) 24 hours post-invasion. A prominent type I interferon response was observed in each cell type, reflecting a secondary response to secreted cytokine in infected cultures. To identify a core cytokine-independent response in T. cruzi-infected fibroblasts and endothelial cells transwell plates were used to distinguish cytokine-dependent and -independent gene expression profiles. This approach revealed the induction of metabolic and signaling pathways involved in cell proliferation, amino acid catabolism and response to wounding as common themes in T. cruzi-infected cells. In addition, the downregulation of genes involved in mitotic cell cycle and cell division predicted that T. cruzi infection may impede host cell cycle progression. The observation of impaired cytokinesis in T. cruzi-infected cells, following nuclear replication, confirmed this prediction.
Conclusion: Metabolic pathways and cellular processes were identified as significantly altered at the transcriptional level in response to T. cruzi infection in a cytokine-independent manner. Several of these alterations are supported by previous studies of T. cruzi metabolic requirements or effects on the host. However, our methods also revealed a T. cruzi-dependent block in the host cell cycle, at the level of cytokinesis, previously unrecognized for this pathogen-host cell interaction.
Figures


Similar articles
-
Modulation of inflammatory response and parasitism by 15-Deoxy-Δ(12,14) prostaglandin J(2) in Trypanosoma cruzi-infected cardiomyocytes.Int J Parasitol. 2011 Apr;41(5):553-62. doi: 10.1016/j.ijpara.2010.12.002. Epub 2011 Jan 6. Int J Parasitol. 2011. PMID: 21215746
-
Mammalian Target of Rapamycin Inhibition in Trypanosoma cruzi-Infected Macrophages Leads to an Intracellular Profile That Is Detrimental for Infection.Front Immunol. 2018 Feb 20;9:313. doi: 10.3389/fimmu.2018.00313. eCollection 2018. Front Immunol. 2018. PMID: 29515594 Free PMC article.
-
Dogs infected with the blood trypomastigote form of Trypanosoma cruzi display an increase expression of cytokines and chemokines plus an intense cardiac parasitism during acute infection.Mol Immunol. 2014 Mar;58(1):92-7. doi: 10.1016/j.molimm.2013.11.007. Epub 2013 Dec 7. Mol Immunol. 2014. PMID: 24317279
-
Understanding Host-Pathogen Interactions in Congenital Chagas Disease Through Transcriptomic Approaches.Pathogens. 2025 Jan 22;14(2):106. doi: 10.3390/pathogens14020106. Pathogens. 2025. PMID: 40005483 Free PMC article. Review.
-
Mechanisms of Trypanosoma cruzi persistence in Chagas disease.Cell Microbiol. 2012 May;14(5):634-43. doi: 10.1111/j.1462-5822.2012.01764.x. Epub 2012 Feb 24. Cell Microbiol. 2012. PMID: 22309180 Free PMC article. Review.
Cited by
-
Trypanosoma cruzi infection induces DNA double-strand breaks and activates DNA damage response pathway in host epithelial cells.Sci Rep. 2024 Mar 4;14(1):5225. doi: 10.1038/s41598-024-53589-w. Sci Rep. 2024. PMID: 38433244 Free PMC article.
-
Early Trypanosoma cruzi infection reprograms human epithelial cells.Biomed Res Int. 2014;2014:439501. doi: 10.1155/2014/439501. Epub 2014 Apr 9. Biomed Res Int. 2014. PMID: 24812617 Free PMC article.
-
Paracrine Signaling Mediated by the Cytosolic Tryparedoxin Peroxidase of Trypanosoma cruzi.Pathogens. 2024 Jan 10;13(1):67. doi: 10.3390/pathogens13010067. Pathogens. 2024. PMID: 38251374 Free PMC article.
-
Trypanosoma cruzi Infection Induces Cellular Stress Response and Senescence-Like Phenotype in Murine Fibroblasts.Front Immunol. 2018 Jul 9;9:1569. doi: 10.3389/fimmu.2018.01569. eCollection 2018. Front Immunol. 2018. PMID: 30038622 Free PMC article.
-
DNA lesions and repair in trypanosomatids infection.Genet Mol Biol. 2020 Mar 27;43(1 suppl. 1):e20190163. doi: 10.1590/1678-4685-GMB-2019-0163. eCollection 2020. Genet Mol Biol. 2020. PMID: 32236391 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

