Measure of synonymous codon usage diversity among genes in bacteria
- PMID: 19480720
- PMCID: PMC2697163
- DOI: 10.1186/1471-2105-10-167
Measure of synonymous codon usage diversity among genes in bacteria
Abstract
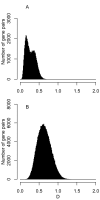
Background: In many bacteria, intragenomic diversity in synonymous codon usage among genes has been reported. However, no quantitative attempt has been made to compare the diversity levels among different genomes. Here, we introduce a mean dissimilarity-based index (Dmean) for quantifying the level of diversity in synonymous codon usage among all genes within a genome.
Results: The application of Dmean to 268 bacterial genomes shows that in bacteria with extremely biased genomic G+C compositions there is little diversity in synonymous codon usage among genes. Furthermore, our findings contradict previous reports. For example, a low level of diversity in codon usage among genes has been reported for Helicobacter pylori, but based on Dmean, the diversity level of this species is higher than those of more than half of bacteria tested here. The discrepancies between our findings and previous reports are probably due to differences in the methods used for measuring codon usage diversity.
Conclusion: We recommend that Dmean be used to measure the diversity level of codon usage among genes. This measure can be applied to other compositional features such as amino acid usage and dinucleotide relative abundance as a genomic signature.
Figures
References
-
- Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988;16:8207–8211. doi: 10.1093/nar/16.17.8207. - DOI - PMC - PubMed
-
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

