A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay
- PMID: 19481524
- PMCID: PMC2712825
- DOI: 10.1016/j.molcel.2009.04.017
A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay
Abstract
Eukaryotes possess numerous quality control systems that monitor both the synthesis of RNA and the integrity of the finished products. We previously demonstrated that Saccharomyces cerevisiae possesses a quality control mechanism, nonfunctional rRNA decay (NRD), capable of detecting and eliminating translationally defective rRNAs. Here we show that NRD can be divided into two mechanistically distinct pathways: one that eliminates rRNAs with deleterious mutations in the decoding site (18S NRD) and one that eliminates rRNAs containing deleterious mutations in the peptidyl transferase center (25S NRD). 18S NRD is dependent on translation elongation and utilizes the same proteins as those participating in no-go mRNA decay (NGD). In cells that accumulate 18S NRD and NGD decay intermediates, both RNA types can be seen in P-bodies. We propose that 18S NRD and NGD are different observable outcomes of the same initiating event: a ribosome stalled inappropriately at a sense codon during translation elongation.
Figures
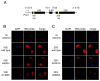


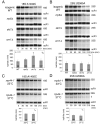
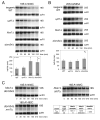
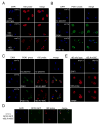

Comment in
-
Related mechanisms for mRNA and rRNA quality control.Mol Cell. 2009 May 14;34(4):401-2. doi: 10.1016/j.molcel.2009.05.008. Mol Cell. 2009. PMID: 19481519
References
-
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
-
- Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. - PubMed
-
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

