Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii
- PMID: 19481527
- PMCID: PMC3268689
- DOI: 10.1016/j.molcel.2009.04.021
Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii
Abstract
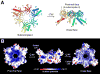
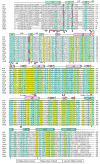
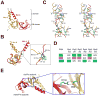
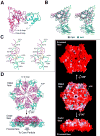
Eukaryotic proteasome consists of a core particle (CP), which degrades unfolded protein, and a regulatory particle (RP), which is responsible for recognition, ATP-dependent unfolding, and translocation of polyubiquitinated substrate protein. In the archaea Methanocaldococcus jannaschii, the RP is a homohexameric complex of proteasome-activating nucleotidase (PAN). Here, we report the crystal structures of essential elements of the archaeal proteasome: the CP, the ATPase domain of PAN, and a distal subcomplex that is likely the first to encounter substrate. The distal subcomplex contains a coiled-coil segment and an OB-fold domain, both of which appear to be conserved in the eukaryotic proteasome. The OB domains of PAN form a hexameric ring with a 13 A pore, which likely constitutes the outermost constriction of the substrate translocation channel. These studies reveal structural codes and architecture of the complete proteasome, identify potential substrate-binding sites, and uncover unexpected asymmetry in the RP of archaea and eukaryotes.
Figures


Comment in
-
The proteasome's crown for destruction.Mol Cell. 2009 Jun 12;34(5):519-20. doi: 10.1016/j.molcel.2009.05.021. Mol Cell. 2009. PMID: 19524532
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta crystallographica. 2002;58:1948–1954. - PubMed
-
- Agrawal V, Kishan KV. OB-fold: growing bigger with functional consistency. Curr Protein Pept Sci. 2003;4:195–206. - PubMed
-
- Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol. 2002;12:794–801. - PubMed
-
- Baumeister W, Lupas A. The proteasome. Current opinion in structural biology. 1997;7:273–278. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

