DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions
- PMID: 19482874
- PMCID: PMC2722982
- DOI: 10.1093/hmg/ddp256
DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions
Abstract
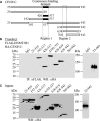
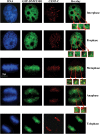
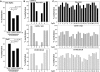
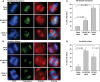
DNA methylation is an epigenetically imposed mark of transcriptional repression that is essential for maintenance of chromatin structure and genomic stability. Genome-wide methylation patterns are mediated by the combined action of three DNA methyltransferases: DNMT1, DNMT3A and DNMT3B. Compelling links exist between DNMT3B and chromosome stability as emphasized by the mitotic defects that are a hallmark of ICF syndrome, a disease arising from germline mutations in DNMT3B. Centromeric and pericentromeric regions are essential for chromosome condensation and the fidelity of segregation. Centromere regions contain distinct epigenetic marks, including dense DNA hypermethylation, yet the mechanisms by which DNA methylation is targeted to these regions remains largely unknown. In the present study, we used a yeast two-hybrid screen and identified a novel interaction between DNMT3B and constitutive centromere protein CENP-C. CENP-C is itself essential for mitosis. We confirm this interaction in mammalian cells and map the domains responsible. Using siRNA knock downs, bisulfite genomic sequencing and ChIP, we demonstrate for the first time that CENP-C recruits DNA methylation and DNMT3B to both centromeric and pericentromeric satellite repeats and that CENP-C and DNMT3B regulate the histone code in these regions, including marks characteristic of centromeric chromatin. Finally, we demonstrate that loss of CENP-C or DNMT3B leads to elevated chromosome misalignment and segregation defects during mitosis and increased transcription of centromeric repeats. Taken together, our data reveal a novel mechanism by which DNA methylation is targeted to discrete regions of the genome and contributes to chromosomal stability.
Figures




Similar articles
-
Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading.Mol Cell. 2012 May 11;46(3):260-73. doi: 10.1016/j.molcel.2012.03.021. Epub 2012 Apr 17. Mol Cell. 2012. PMID: 22516971
-
DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development.Cell. 1999 Oct 29;99(3):247-57. doi: 10.1016/s0092-8674(00)81656-6. Cell. 1999. PMID: 10555141
-
Budding yeast CENP-ACse4 interacts with the N-terminus of Sgo1 and regulates its association with centromeric chromatin.Cell Cycle. 2018;17(1):11-23. doi: 10.1080/15384101.2017.1380129. Epub 2018 Jan 2. Cell Cycle. 2018. PMID: 28980861 Free PMC article.
-
The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres.Chromosome Res. 2004;12(6):543-56. doi: 10.1023/B:CHRO.0000036593.72788.99. Chromosome Res. 2004. PMID: 15289662 Review.
-
Variations on a nucleosome theme: The structural basis of centromere function.Bioessays. 2017 Apr;39(4). doi: 10.1002/bies.201600241. Epub 2017 Feb 21. Bioessays. 2017. PMID: 28220502 Review.
Cited by
-
DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function.Dev Cell. 2015 May 4;33(3):314-27. doi: 10.1016/j.devcel.2015.03.020. Dev Cell. 2015. PMID: 25942623 Free PMC article.
-
The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation.J Biol Chem. 2010 Aug 20;285(34):26114-20. doi: 10.1074/jbc.M109.089433. Epub 2010 Jun 11. J Biol Chem. 2010. PMID: 20547484 Free PMC article.
-
Epigenetic regulation of centromere function.Cell Mol Life Sci. 2020 Aug;77(15):2899-2917. doi: 10.1007/s00018-020-03460-8. Epub 2020 Feb 1. Cell Mol Life Sci. 2020. PMID: 32008088 Free PMC article. Review.
-
Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres.Plant Cell. 2013 Sep;25(9):3389-404. doi: 10.1105/tpc.113.114736. Epub 2013 Sep 6. Plant Cell. 2013. PMID: 24014547 Free PMC article.
-
Emerging Roles for Transcription Factors During Mitosis.Cells. 2025 Feb 12;14(4):263. doi: 10.3390/cells14040263. Cells. 2025. PMID: 39996736 Free PMC article. Review.
References
-
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. - PubMed
-
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature Rev. Genet. 2002;3:662–673. - PubMed
-
- Robertson K.D. DNA methylation and human disease. Nature Rev. Genet. 2005;6:597–610. - PubMed
-
- Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nature Rev. Genet. 2002;3:415–428. - PubMed
-
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

