Steric gate variants of UmuC confer UV hypersensitivity on Escherichia coli
- PMID: 19482923
- PMCID: PMC2715726
- DOI: 10.1128/JB.01742-08
Steric gate variants of UmuC confer UV hypersensitivity on Escherichia coli
Abstract
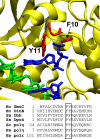
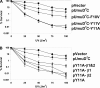
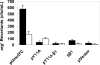
Y family DNA polymerases are specialized for replication of damaged DNA and represent a major contribution to cellular resistance to DNA lesions. Although the Y family polymerase active sites have fewer contacts with their DNA substrates than replicative DNA polymerases, Y family polymerases appear to exhibit specificity for certain lesions. Thus, mutation of the steric gate residue of Escherichia coli DinB resulted in the specific loss of lesion bypass activity. We constructed variants of E. coli UmuC with mutations of the steric gate residue Y11 and of residue F10 and determined that strains harboring these variants are hypersensitive to UV light. Moreover, these UmuC variants are dominant negative with respect to sensitivity to UV light. The UV hypersensitivity and the dominant negative phenotype are partially suppressed by additional mutations in the known motifs in UmuC responsible for binding to the beta processivity clamp, suggesting that the UmuC steric gate variant exerts its effects via access to the replication fork. Strains expressing the UmuC Y11A variant also exhibit decreased UV mutagenesis. Strikingly, disruption of the dnaQ gene encoding the replicative DNA polymerase proofreading subunit suppressed the dominant negative phenotype of a UmuC steric gate variant. This could be due to a recruitment function of the proofreading subunit or involvement of the proofreading subunit in a futile cycle of base insertion/excision with the UmuC steric gate variant.
Figures





Similar articles
-
Characterization of Escherichia coli UmuC active-site loops identifies variants that confer UV hypersensitivity.J Bacteriol. 2011 Oct;193(19):5400-11. doi: 10.1128/JB.05301-11. Epub 2011 Jul 22. J Bacteriol. 2011. PMID: 21784925 Free PMC article.
-
Point mutations in Escherichia coli DNA pol V that confer resistance to non-cognate DNA damage also alter protein-protein interactions.Mutat Res. 2015 Oct;780:1-14. doi: 10.1016/j.mrfmmm.2015.07.003. Epub 2015 Jul 13. Mutat Res. 2015. PMID: 26218456
-
Characterization of novel alleles of the Escherichia coli umuDC genes identifies additional interaction sites of UmuC with the beta clamp.J Bacteriol. 2009 Oct;191(19):5910-20. doi: 10.1128/JB.00292-09. Epub 2009 Jul 24. J Bacteriol. 2009. PMID: 19633075 Free PMC article.
-
Y-family DNA polymerases in Escherichia coli.Trends Microbiol. 2007 Feb;15(2):70-7. doi: 10.1016/j.tim.2006.12.004. Epub 2007 Jan 4. Trends Microbiol. 2007. PMID: 17207624 Review.
-
Current understanding of UV-induced base pair substitution mutation in E. coli with particular reference to the DNA polymerase III complex.Mutat Res. 1987 Dec;181(2):219-26. doi: 10.1016/0027-5107(87)90099-6. Mutat Res. 1987. PMID: 3317025 Review.
Cited by
-
Characterization of Escherichia coli UmuC active-site loops identifies variants that confer UV hypersensitivity.J Bacteriol. 2011 Oct;193(19):5400-11. doi: 10.1128/JB.05301-11. Epub 2011 Jul 22. J Bacteriol. 2011. PMID: 21784925 Free PMC article.
-
Escherichia coli UmuC active site mutants: effects on translesion DNA synthesis, mutagenesis and cell survival.DNA Repair (Amst). 2012 Sep 1;11(9):726-32. doi: 10.1016/j.dnarep.2012.06.005. Epub 2012 Jul 10. DNA Repair (Amst). 2012. PMID: 22784977 Free PMC article.
-
UV-B-Induced DNA Repair Mechanisms and Their Effects on Mutagenesis and Culturability in Escherichia coli.bioRxiv [Preprint]. 2025 Jun 9:2024.11.14.623584. doi: 10.1101/2024.11.14.623584. bioRxiv. 2025. PMID: 39605428 Free PMC article. Preprint.
-
Steric gate residues of Y-family DNA polymerases DinB and pol kappa are crucial for dNTP-induced conformational change.DNA Repair (Amst). 2015 May;29:65-73. doi: 10.1016/j.dnarep.2015.01.012. Epub 2015 Feb 4. DNA Repair (Amst). 2015. PMID: 25684709 Free PMC article.
-
Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity.Nucleic Acids Res. 2012 Jul;40(13):6144-57. doi: 10.1093/nar/gks233. Epub 2012 Mar 15. Nucleic Acids Res. 2012. PMID: 22422840 Free PMC article.
References
-
- Becherel, O. J., R. P. P. Fuchs, and J. Wagner. 2002. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair 1703-708. - PubMed
-
- Beuning, P. J., D. Sawicka, D. Barsky, and G. C. Walker. 2006. Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59460-474. - PubMed
-
- Beuning, P. J., S. M. Simon, V. G. Godoy, D. F. Jarosz, and G. C. Walker. 2006. Characterization of Escherichia coli translesion synthesis polymerases and their accessory factors. Methods Enzymol. 408318-340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

