Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs
- PMID: 19482942
- PMCID: PMC2688437
- DOI: 10.1073/pnas.0901997106
Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs
Abstract
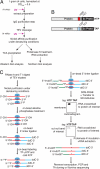
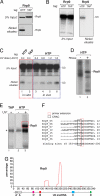
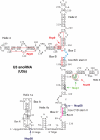
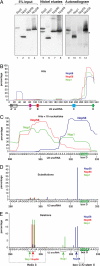
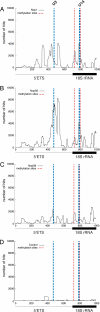
The U3 small nucleolar ribonucleoprotein (snoRNP) plays an essential role in ribosome biogenesis but, like many RNA-protein complexes, its architecture is poorly understood. To address this problem, binding sites for the snoRNP proteins Nop1, Nop56, Nop58, and Rrp9 were mapped by UV cross-linking and analysis of cDNAs. Cross-linked protein-RNA complexes were purified under highly-denaturing conditions, ensuring that only direct interactions were detected. Recovered RNA fragments were amplified after linker ligation and cDNA synthesis. Cross-linking was successfully performed either in vitro on purified complexes or in vivo in living cells. Cross-linking sites were precisely mapped either by Sanger sequencing of multiple cloned fragments or direct, high-throughput Solexa sequencing. Analysis of RNAs associated with the snoRNP proteins revealed remarkably high signal-to-noise ratios and identified specific binding sites for each of these proteins on the U3 RNA. The results were consistent with previous data, demonstrating the reliability of the method, but also provided insights into the architecture of the U3 snoRNP. The snoRNP proteins were also cross-linked to pre-rRNA fragments, with preferential association at known sites of box C/D snoRNA function. This finding demonstrates that the snoRNP proteins directly contact the pre-rRNA substrate, suggesting roles in snoRNA recruitment. The techniques reported here should be widely applicable to analyses of RNA-protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. - PubMed
-
- Gilbert C, Svejstrup JQ. Ausubel FM, editor. RNA immunoprecipitation for determining RNA-protein associations in vivo. Current Protocols in Molecular Biology. 2006:27.4.1–27.4.11. Chapter 27. - PubMed
-
- Urlaub H, Hartmuth K, Kostka S, Grelle G, Luhrmann R. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA-protein contacts in native U1 and U4/U6.U5 snRNPs. J Biol Chem. 2000;275:41458–41468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

