Lin28 promotes transformation and is associated with advanced human malignancies
- PMID: 19483683
- PMCID: PMC2757943
- DOI: 10.1038/ng.392
Lin28 promotes transformation and is associated with advanced human malignancies
Abstract
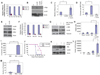
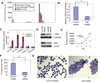
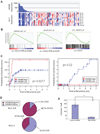
Multiple members of the let-7 family of miRNAs are often repressed in human cancers, thereby promoting oncogenesis by derepressing targets such as HMGA2, K-Ras and c-Myc. However, the mechanism by which let-7 miRNAs are coordinately repressed is unclear. The RNA-binding proteins LIN28 and LIN28B block let-7 precursors from being processed to mature miRNAs, suggesting that their overexpression might promote malignancy through repression of let-7. Here we show that LIN28 and LIN28B are overexpressed in primary human tumors and human cancer cell lines (overall frequency approximately 15%), and that overexpression is linked to repression of let-7 family miRNAs and derepression of let-7 targets. LIN28 and LIN28b facilitate cellular transformation in vitro, and overexpression is associated with advanced disease across multiple tumor types. Our work provides a mechanism for the coordinate repression of let-7 miRNAs observed in a subset of human cancers, and associates activation of LIN28 and LIN28B with poor clinical prognosis.
Figures
Comment in
-
Tumors line up for a letdown.Nat Genet. 2009 Jul;41(7):768-9. doi: 10.1038/ng0709-768. Nat Genet. 2009. PMID: 19557079
-
A Yin-Yang balancing act of the lin28/let-7 link in tumorigenesis.J Hepatol. 2010 Nov;53(5):974-5. doi: 10.1016/j.jhep.2010.07.001. Epub 2010 Jul 21. J Hepatol. 2010. PMID: 20739081 Free PMC article. No abstract available.
References
-
- Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. - PubMed
-
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat.Genet. 2007;39:673–677. - PubMed
-
- Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol.Cell. 2008;32:276–284. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

