Protein interaction platforms: visualization of interacting proteins in yeast
- PMID: 19483691
- PMCID: PMC2819136
- DOI: 10.1038/nmeth.1337
Protein interaction platforms: visualization of interacting proteins in yeast
Abstract
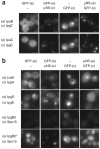
Here we describe the protein interaction platform assay, a method for identifying interacting proteins in Saccharomyces cerevisiae. This assay relies on the reovirus scaffolding protein microNS, which forms large focal inclusions in living cells. When a query protein is fused to microNS and potential interaction partners are fused to a fluorescent reporter, interactors can be identified by screening for yeast that display fluorescent foci.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

