Quantifying biogenic bias in screening libraries
- PMID: 19483698
- PMCID: PMC2783405
- DOI: 10.1038/nchembio.180
Quantifying biogenic bias in screening libraries
Abstract
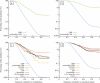
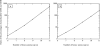
In lead discovery, libraries of 10(6) molecules are screened for biological activity. Given the over 10(60) drug-like molecules thought possible, such screens might never succeed. The fact that they do, even occasionally, implies a biased selection of library molecules. We have developed a method to quantify the bias in screening libraries toward biogenic molecules. With this approach, we consider what is missing from screening libraries and how they can be optimized.
Figures


References
-
- Wilhelm S, et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat. Rev. Drug Discovery. 2006;5:835–844. - PubMed
-
- Spencer RW. High-throughput screening of historic collections: observations on file size, biological targets, and file diversity. Biotechnol. Bioeng. 1998;61:61–67. - PubMed
-
- Fox S, Farr-Jones S, Sopchak L, Boggs A, Comley J. High-Throughput Screening: Searching for Higher Productivity. J. Biomol. Screen. 2004;9:354–358. - PubMed
-
- Macarron R. Critical review of the role of HTS in drug discovery. Drug Discov. Today. 2006;11:277–279. - PubMed
Publication types
MeSH terms
Substances
Associated data
- PubChem-Substance/58097293
- PubChem-Substance/58097294
- PubChem-Substance/58097295
- PubChem-Substance/58097296
- PubChem-Substance/58097297
- PubChem-Substance/58097298
- PubChem-Substance/58097299
- PubChem-Substance/58097300
- PubChem-Substance/58097301
- PubChem-Substance/58097302
- PubChem-Substance/58097303
- PubChem-Substance/58097304
- PubChem-Substance/58097305
- PubChem-Substance/58097306
- PubChem-Substance/58097307
- PubChem-Substance/58097308
- PubChem-Substance/58097309
- PubChem-Substance/58097310
- PubChem-Substance/58097311
- PubChem-Substance/58097312
- PubChem-Substance/58097313
- PubChem-Substance/58097314
- PubChem-Substance/58097315
- PubChem-Substance/58097316
- PubChem-Substance/58097317
- PubChem-Substance/58097318
- PubChem-Substance/58097319
- PubChem-Substance/58097320
- PubChem-Substance/58097321
- PubChem-Substance/58097322
- PubChem-Substance/58097323
- PubChem-Substance/58097324
- PubChem-Substance/58097325
- PubChem-Substance/58097326
- PubChem-Substance/58097327
- PubChem-Substance/58097328
- PubChem-Substance/58097329
- PubChem-Substance/58097330
- PubChem-Substance/58097331
- PubChem-Substance/58097332
- PubChem-Substance/58097333
- PubChem-Substance/58097334
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

