Anchorage-independent cell growth signature identifies tumors with metastatic potential
- PMID: 19483725
- PMCID: PMC3008357
- DOI: 10.1038/onc.2009.139
Anchorage-independent cell growth signature identifies tumors with metastatic potential
Abstract
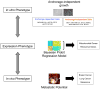
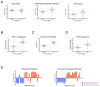
The oncogenic phenotype is complex, resulting from the accumulation of multiple somatic mutations that lead to the deregulation of growth regulatory and cell fate controlling activities and pathways. The ability to dissect this complexity, so as to reveal discrete aspects of the biology underlying the oncogenic phenotype, is critical to understanding the various mechanisms of disease as well as to reveal opportunities for novel therapeutic strategies. Previous work has characterized the process of anchorage-independent growth of cancer cells in vitro as a key aspect of the tumor phenotype, particularly with respect to metastatic potential. Nevertheless, it remains a major challenge to translate these cell biology findings into the context of human tumors. We previously used DNA microarray assays to develop expression signatures, which have the capacity to identify subtle distinctions in biological states and can be used to connect in vitro and in vivo states. Here we describe the development of a signature of anchorage-independent growth, show that the signature exhibits characteristics of deregulated mitochondrial function and then demonstrate that the signature identifies human tumors with the potential for metastasis.
Figures



References
-
- Bild A, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi M-B, Harpole D, Lancaster JM, Berchuck A, Olson JA, Marks JR, Dressman HK, West M, Nevins JR. Nature. 2006;439:353–357. - PubMed
-
- Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Nat Genet. 2006;38:576–82. - PubMed
-
- Campbell PM, Der CJ. Semin Cancer Biol. 2004;14:105–14. - PubMed
-
- Chang JT, Nevins JR. Bioinformatics. 2006;22:2926–2933. - PubMed
-
- Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, 3rd, Felding-Habermann B. Cancer Res. 2007;67:1472–86. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

