A CA(+) pair adjacent to a sheared GA or AA pair stabilizes size-symmetric RNA internal loops
- PMID: 19485416
- PMCID: PMC2697601
- DOI: 10.1021/bi8019405
A CA(+) pair adjacent to a sheared GA or AA pair stabilizes size-symmetric RNA internal loops
Abstract
RNA internal loops are often important sites for folding and function. Residues in internal loops can have pKa values shifted close to neutral pH because of the local structural environment. A series of RNA internal loops were studied at different pH by UV absorbance versus temperature melting experiments and imino proton nuclear magnetic resonance (NMR). A stabilizing CA pair forms at pH 7 in the CG/AA and CA/AA nearest neighbors when the CA pair is the first noncanonical pair (loop-terminal pair) in 3 x 3 nucleotide and larger size-symmetric internal loops. These CG/AA and CA/AA nearest neighbors, with CA adjacent to a closing Watson-Crick pair, are further stabilized when the pH is lowered from 7 to 5.5. The results are consistent with a significantly larger fraction (from approximately 20% at pH 7 to approximately 90% at pH 5.5) of adenines being protonated at the N1 position to form stabilizing wobble CA+ pairs adjacent to a sheared GA or AA pair. The noncanonical pair adjacent to the GA pair in CG/AA can either stabilize or destabilize the loop, consistent with the sequence-dependent thermodynamics of GA pairs. No significant pH-dependent stabilization is found for most of the other nearest neighbor combinations involving CA pairs (e.g., CA/AG and AG/CA), which is consistent with the formation of various nonwobble pairs observed in different local sequence contexts in crystal and NMR structures. A revised free-energy model, including stabilization by wobble CA+ pairs, is derived for predicting stabilities of medium-size RNA internal loops.
Figures
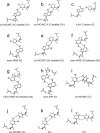

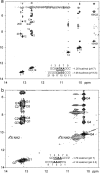
 (CT = 0.5 mM, pH 5.3, 0 °C, see Figure 2d for 1D spectrum). There is a very weak cross-peak of G1H1−G19H1 (not shown). The imino protons of G5, G14, and G15 have chemical shifts and cross-peaks typical of consecutive sheared GA pairs (16,20,72). The G15 amino protons resonate at 9.2 and 5.5 ppm, respectively, suggesting the formation of sheared GA pairs with G5 and G15 in the C2′-endo sugar pucker (73,74). There is no indication of the formation of A+C pair in this loop. (b)
(CT = 0.5 mM, pH 5.3, 0 °C, see Figure 2d for 1D spectrum). There is a very weak cross-peak of G1H1−G19H1 (not shown). The imino protons of G5, G14, and G15 have chemical shifts and cross-peaks typical of consecutive sheared GA pairs (16,20,72). The G15 amino protons resonate at 9.2 and 5.5 ppm, respectively, suggesting the formation of sheared GA pairs with G5 and G15 in the C2′-endo sugar pucker (73,74). There is no indication of the formation of A+C pair in this loop. (b)  (CT = 1.5 mM, pH 5.1, −5 °C, see Figure 2f for 1D spectrum). The cross-peak of G1H1−G7H1 is unresolved because of overlap but is observed in
(CT = 1.5 mM, pH 5.1, −5 °C, see Figure 2f for 1D spectrum). The cross-peak of G1H1−G7H1 is unresolved because of overlap but is observed in  (see and Figure 2e for 1D spectrum). The broad peak at ∼10.6 ppm is likely due to the amino protons of A+6, which shows a strong cross-peak to the other amino proton and a weak cross-peak to the G7 imino proton. Adenine amino protons with similar chemical shift have been observed in other cases of CA+ pairs (12). The G4 amino protons resonate at 8.8 and 6.2 ppm, respectively, suggesting the formation of sheared GA pairs with G4 in the C2′-endo sugar pucker (73,74).
(see and Figure 2e for 1D spectrum). The broad peak at ∼10.6 ppm is likely due to the amino protons of A+6, which shows a strong cross-peak to the other amino proton and a weak cross-peak to the G7 imino proton. Adenine amino protons with similar chemical shift have been observed in other cases of CA+ pairs (12). The G4 amino protons resonate at 8.8 and 6.2 ppm, respectively, suggesting the formation of sheared GA pairs with G4 in the C2′-endo sugar pucker (73,74).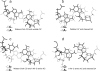
 segment of the NMR structure of the substrate loop of VS ribozyme (9). The Watson−Crick CG, A N1−C amino single hydrogen-bond AC pair, and sheared GA shown in c and d are taken from
segment of the NMR structure of the substrate loop of VS ribozyme (9). The Watson−Crick CG, A N1−C amino single hydrogen-bond AC pair, and sheared GA shown in c and d are taken from  segment of the NMR structure of loop B of a hairpin ribozyme (37). The stacking figures are generated by the 3DNA program (75).
segment of the NMR structure of loop B of a hairpin ribozyme (37). The stacking figures are generated by the 3DNA program (75).References
-
- Legault P.; Pardi A. (1997) Unusual dynamics and pKa shift at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 119, 6621–6628.
-
- Bevilacqua P. C. (2003) Mechanistic considerations for general acid−base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry 42, 2259–2265. - PubMed
-
- Cai Z.; Tinoco I. Jr. (1996) Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35, 6026–6036. - PubMed
-
- Allawi H. T.; SantaLucia J. Jr. (1998) Nearest-neighbor thermodynamics of internal AC mismatches in DNA: Sequence dependence and pH effects. Biochemistry 37, 9435–9444. - PubMed
-
- SantaLucia J. Jr.; Kierzek R.; Turner D. H. (1991) Stabilities of consecutive A·C, C·C, G·G, U·C, and U·U mismatches in RNA internal loops: Evidence for stable hydrogen bonded U·U and C·C+ pairs. Biochemistry 30, 8242–8251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

