Differential expression of utrophin-A and -B promoters in the central nervous system (CNS) of normal and dystrophic mdx mice
- PMID: 19486009
- PMCID: PMC8095546
- DOI: 10.1111/j.1750-3639.2009.00275.x
Differential expression of utrophin-A and -B promoters in the central nervous system (CNS) of normal and dystrophic mdx mice
Abstract
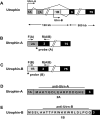
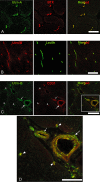
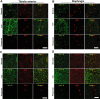
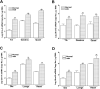
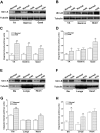
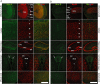
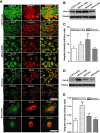
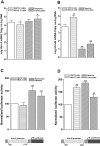
Utrophin (Utrn) is the autosomal homolog of dystrophin, the Duchene Muscular Dystrophy (DMD) locus product and of therapeutic interest, as its overexpression can compensate dystrophin's absence. Utrn is transcribed by Utrn-A and -B promoters with mRNAs differing at their 5' ends. However, previous central nervous system (CNS) studies used C-terminal antibodies recognizing both isoforms. As this distinction may impact upregulation strategies, we generated Utrn-A and -B promoter-specific antibodies, Taqman Polymerase chain reaction (PCR)-based absolute copy number assays, and luciferase-reporter constructs to study CNS of normal and dystrophic mdx mice. Differential expression of Utrn-A and -B was noted in microdissected and capillary-enriched fractions. At the protein level, Utrn-B was predominantly expressed in vasculature and ependymal lining, whereas Utrn-A was expressed in neurons, astrocytes, choroid plexus and pia mater. mRNA quantification demonstrated matching patterns of differential expression; however, transcription-translation mismatch was noted for Utrn-B in caudal brain regions. Utrn-A and Utrn-B proteins were significantly upregulated in olfactory bulb and cerebellum of mdx brain. Differential promoter activity, mRNA and protein expressions were studied in cultured C2C12, bEnd3, neurons and astrocytes. Promoter activity ranking for Utrn-A and -B was neurons > astrocytes > C2C12 > bEnd3 and bEnd3 > astrocytes > neurons > C2C12, respectively. Our results identify promoter usage patterns for therapeutic targeting and define promoter-specific differential distribution of Utrn isoforms in normal and dystrophic CNS.
Figures



Similar articles
-
PMO-based let-7c site blocking oligonucleotide (SBO) mediated utrophin upregulation in mdx mice, a therapeutic approach for Duchenne muscular dystrophy (DMD).Sci Rep. 2020 Dec 9;10(1):21492. doi: 10.1038/s41598-020-76338-1. Sci Rep. 2020. PMID: 33298994 Free PMC article.
-
Comparison of skeletal muscle pathology and motor function of dystrophin and utrophin deficient mouse strains.Neuromuscul Disord. 2012 May;22(5):406-17. doi: 10.1016/j.nmd.2011.10.011. Epub 2012 Jan 27. Neuromuscul Disord. 2012. PMID: 22284942
-
Utrophin haploinsufficiency does not worsen the functional performance, resistance to eccentric contractions and force production of dystrophic mice.PLoS One. 2018 Jun 7;13(6):e0198408. doi: 10.1371/journal.pone.0198408. eCollection 2018. PLoS One. 2018. PMID: 29879154 Free PMC article.
-
Haploinsufficiency of utrophin gene worsens skeletal muscle inflammation and fibrosis in mdx mice.J Neurol Sci. 2008 Jan 15;264(1-2):106-11. doi: 10.1016/j.jns.2007.08.029. Epub 2007 Sep 24. J Neurol Sci. 2008. PMID: 17889902 Free PMC article.
-
Lifelong quercetin enrichment and cardioprotection in Mdx/Utrn+/- mice.Am J Physiol Heart Circ Physiol. 2017 Jan 1;312(1):H128-H140. doi: 10.1152/ajpheart.00552.2016. Epub 2016 Nov 11. Am J Physiol Heart Circ Physiol. 2017. PMID: 27836895
Cited by
-
Identifying FDA-Approved Drugs that Upregulate Utrophin A as a Therapeutic Strategy for Duchenne Muscular Dystrophy.Methods Mol Biol. 2023;2587:495-510. doi: 10.1007/978-1-0716-2772-3_26. Methods Mol Biol. 2023. PMID: 36401046
-
Dystrophins, utrophins, and associated scaffolding complexes: role in mammalian brain and implications for therapeutic strategies.J Biomed Biotechnol. 2010;2010:849426. doi: 10.1155/2010/849426. Epub 2010 Jun 17. J Biomed Biotechnol. 2010. PMID: 20625423 Free PMC article. Review.
-
Drug discovery for Duchenne muscular dystrophy via utrophin promoter activation screening.PLoS One. 2011;6(10):e26169. doi: 10.1371/journal.pone.0026169. Epub 2011 Oct 20. PLoS One. 2011. PMID: 22028826 Free PMC article.
-
PMO-based let-7c site blocking oligonucleotide (SBO) mediated utrophin upregulation in mdx mice, a therapeutic approach for Duchenne muscular dystrophy (DMD).Sci Rep. 2020 Dec 9;10(1):21492. doi: 10.1038/s41598-020-76338-1. Sci Rep. 2020. PMID: 33298994 Free PMC article.
-
Targeting IRES-dependent translation as a novel approach for treating Duchenne muscular dystrophy.RNA Biol. 2021 Sep;18(9):1238-1251. doi: 10.1080/15476286.2020.1847894. Epub 2020 Nov 19. RNA Biol. 2021. PMID: 33164678 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

