Functional roles of the pre-sensor I insertion sequence in an AAA+ bacterial enhancer binding protein
- PMID: 19486295
- PMCID: PMC2745333
- DOI: 10.1111/j.1365-2958.2009.06744.x
Functional roles of the pre-sensor I insertion sequence in an AAA+ bacterial enhancer binding protein
Abstract
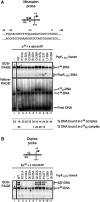
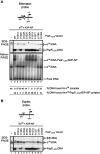
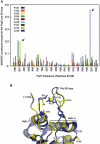
Molecular machines belonging to the AAA+ superfamily of ATPases use NTP hydrolysis to remodel their versatile substrates. The presence of an insertion sequence defines the major phylogenetic pre-sensor I insertion (pre-SIi) AAA+ superclade. In the bacterial sigma(54)-dependent enhancer binding protein phage shock protein F (PspF) the pre-SIi loop adopts different conformations depending on the nucleotide-bound state. Single amino acid substitutions within the dynamic pre-SIi loop of PspF drastically change the ATP hydrolysis parameters, indicating a structural link to the distant hydrolysis site. We used a site-specific protein-DNA proximity assay to measure the contribution of the pre-SIi loop in sigma(54)-dependent transcription and demonstrate that the pre-SIi loop is a major structural feature mediating nucleotide state-dependent differential engagement with Esigma(54). We suggest that much, if not all, of the action of the pre-SIi loop is mediated through the L1 loop and relies on a conserved molecular switch, identified in a crystal structure of one pre-SIi variant and in accordance with the high covariance between some pre-SIi residues and distinct residues outside the pre-SIi sequence.
Figures



Similar articles
-
Sensor I threonine of the AAA+ ATPase transcriptional activator PspF is involved in coupling nucleotide triphosphate hydrolysis to the restructuring of sigma 54-RNA polymerase.J Biol Chem. 2007 Mar 30;282(13):9825-9833. doi: 10.1074/jbc.M611532200. Epub 2007 Jan 22. J Biol Chem. 2007. PMID: 17242399
-
The ATP hydrolyzing transcription activator phage shock protein F of Escherichia coli: identifying a surface that binds sigma 54.Proc Natl Acad Sci U S A. 2003 Mar 4;100(5):2278-83. doi: 10.1073/pnas.0537525100. Epub 2003 Feb 24. Proc Natl Acad Sci U S A. 2003. PMID: 12601152 Free PMC article.
-
A key hydrophobic patch identified in an AAA⁺ protein essential for its in trans inhibitory regulation.J Mol Biol. 2013 Aug 9;425(15):2656-69. doi: 10.1016/j.jmb.2013.04.024. Epub 2013 May 7. J Mol Biol. 2013. PMID: 23659791 Free PMC article.
-
Investigating protein-protein interfaces in bacterial transcription complexes: a fragmentation approach.Bioessays. 2003 Dec;25(12):1150-3. doi: 10.1002/bies.10388. Bioessays. 2003. PMID: 14635249 Review.
-
Structures and organisation of AAA+ enhancer binding proteins in transcriptional activation.J Struct Biol. 2006 Oct;156(1):190-9. doi: 10.1016/j.jsb.2006.01.006. Epub 2006 Feb 20. J Struct Biol. 2006. PMID: 16531068 Review.
Cited by
-
Rewiring cell signalling through chimaeric regulatory protein engineering.Biochem Soc Trans. 2013 Oct;41(5):1195-200. doi: 10.1042/BST20130138. Biochem Soc Trans. 2013. PMID: 24059508 Free PMC article. Review.
-
Cryo-EM structures of human ClpXP reveal mechanisms of assembly and proteolytic activation.bioRxiv [Preprint]. 2024 Nov 13:2024.11.12.623337. doi: 10.1101/2024.11.12.623337. bioRxiv. 2024. PMID: 39605471 Free PMC article. Preprint.
-
Analyses of dynein heavy chain mutations reveal complex interactions between dynein motor domains and cellular dynein functions.Genetics. 2012 Aug;191(4):1157-79. doi: 10.1534/genetics.112.141580. Epub 2012 May 29. Genetics. 2012. PMID: 22649085 Free PMC article.
-
Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity.Nat Commun. 2011 Feb 1;2:177. doi: 10.1038/ncomms1177. Nat Commun. 2011. PMID: 21285955 Free PMC article.
-
Engineered interfaces of an AAA+ ATPase reveal a new nucleotide-dependent coordination mechanism.J Biol Chem. 2010 May 14;285(20):15178-15186. doi: 10.1074/jbc.M110.103150. Epub 2010 Mar 2. J Biol Chem. 2010. PMID: 20197281 Free PMC article.
References
-
- Burrows PC, Wigneshweraraj SR, Buck M. Protein–DNA interactions that govern AAA+ activator-dependent bacterial transcription initiation. J Mol Biol. 2008;375:43–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

