Symbiotic relationship between brain structure and dynamics
- PMID: 19486538
- PMCID: PMC2700812
- DOI: 10.1186/1471-2202-10-55
Symbiotic relationship between brain structure and dynamics
Abstract
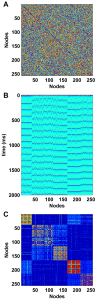
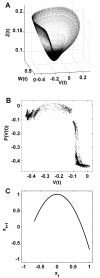
Background: Brain structure and dynamics are interdependent through processes such as activity-dependent neuroplasticity. In this study, we aim to theoretically examine this interdependence in a model of spontaneous cortical activity. To this end, we simulate spontaneous brain dynamics on structural connectivity networks, using coupled nonlinear maps. On slow time scales structural connectivity is gradually adjusted towards the resulting functional patterns via an unsupervised, activity-dependent rewiring rule. The present model has been previously shown to generate cortical-like, modular small-world structural topology from initially random connectivity. We provide further biophysical justification for this model and quantitatively characterize the relationship between structure, function and dynamics that accompanies the ensuing self-organization.
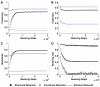
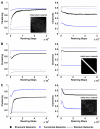
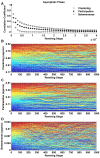
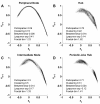
Results: We show that coupled chaotic dynamics generate ordered and modular functional patterns, even on a random underlying structural connectivity. Consequently, structural connectivity becomes more modular as it rewires towards these functional patterns. Functional networks reflect the underlying structural networks on slow time scales, but significantly less so on faster time scales. In spite of ordered functional topology, structural networks remain robustly interconnected--and therefore small-world--due to the presence of central, inter-modular hub nodes. The noisy dynamics of these hubs enable them to persist despite ongoing rewiring and despite their comparative absence in functional networks.
Conclusion: Our results outline a theoretical mechanism by which brain dynamics may facilitate neuroanatomical self-organization. We find time scale dependent differences between structural and functional networks. These differences are likely to arise from the distinct dynamics of central structural nodes.
Figures

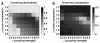


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

