Gene silencing in the marine diatom Phaeodactylum tricornutum
- PMID: 19487243
- PMCID: PMC2724275
- DOI: 10.1093/nar/gkp448
Gene silencing in the marine diatom Phaeodactylum tricornutum
Abstract
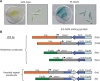
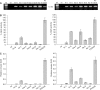
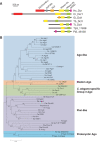
Diatoms are a major but poorly understood phytoplankton group. The recent completion of two whole genome sequences has revealed that they contain unique combinations of genes, likely recruited during their history as secondary endosymbionts, as well as by horizontal gene transfer from bacteria. A major limitation for the study of diatom biology and gene function is the lack of tools to generate targeted gene knockout or knockdown mutants. In this work, we have assessed the possibility of triggering gene silencing in Phaeodactylum tricornutum using constructs containing either anti-sense or inverted repeat sequences of selected target genes. We report the successful silencing of a GUS reporter gene expressed in transgenic lines, as well as the knockdown of endogenous phytochrome (DPH1) and cryptochrome (CPF1) genes. To highlight the utility of the approach we also report the first phenotypic characterization of a diatom mutant (cpf1). Our data open the way for reverse genetics in diatoms and represent a major advance for understanding their biology and ecology. Initial molecular analyses reveal that targeted downregulation likely occurs through transcriptional and post-transcriptional gene silencing mechanisms. Interestingly, molecular players involved in RNA silencing in other eukaryotes are only poorly conserved in diatoms.
Figures



References
-
- Falkowski PG, Barber RT, Smetacek VV. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281:200–207. - PubMed
-
- Smetacek V. Diatoms and the ocean carbon cycle. Protist. 1999;150:25–32. - PubMed
-
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. - PubMed
-
- Montsant A, Andrew EA, Coesel S, De Martino A, Falciatore A, Heijde M, Jabbari K, Maheswari U, Mangogna M, Rayko E, et al. Identification and comparative genomic analysis of signaling and regulatory mechanisms in the diatom Thalassiosira pseudonana. J. Phycol. 2007;43:585–604.
-
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

