Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection
- PMID: 19487424
- PMCID: PMC2715054
- DOI: 10.1084/jem.20090378
Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection
Abstract
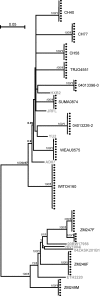
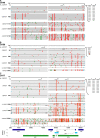
Identification of full-length transmitted HIV-1 genomes could be instrumental in HIV-1 pathogenesis, microbicide, and vaccine research by enabling the direct analysis of those viruses actually responsible for productive clinical infection. We show in 12 acutely infected subjects (9 clade B and 3 clade C) that complete HIV-1 genomes of transmitted/founder viruses can be inferred by single genome amplification and sequencing of plasma virion RNA. This allowed for the molecular cloning and biological analysis of transmitted/founder viruses and a comprehensive genome-wide assessment of the genetic imprint left on the evolving virus quasispecies by a composite of host selection pressures. Transmitted viruses encoded intact canonical genes (gag-pol-vif-vpr-tat-rev-vpu-env-nef) and replicated efficiently in primary human CD4(+) T lymphocytes but much less so in monocyte-derived macrophages. Transmitted viruses were CD4 and CCR5 tropic and demonstrated concealment of coreceptor binding surfaces of the envelope bridging sheet and variable loop 3. 2 mo after infection, transmitted/founder viruses in three subjects were nearly completely replaced by viruses differing at two to five highly selected genomic loci; by 12-20 mo, viruses exhibited concentrated mutations at 17-34 discrete locations. These findings reveal viral properties associated with mucosal HIV-1 transmission and a limited set of rapidly evolving adaptive mutations driven primarily, but not exclusively, by early cytotoxic T cell responses.
Figures




References
-
- Haase A.T. 2005. Perils at mucosal front lines for HIV and SIV and their hosts.Nat. Rev. Immunol. 5:783–792 - PubMed
-
- Margolis L., Shattock R. 2006. Selective transmission of CCR5-utilizing HIV-1: the ‘gatekeeper’ problem resolved? Nat. Rev. Microbiol. 4:312–317 - PubMed
-
- Pope M., Haase A.T. 2003. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection.Nat. Med. 9:847–852 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

