Cyclin-dependent kinase inhibits reinitiation of a normal S-phase program during G2 in fission yeast
- PMID: 19487461
- PMCID: PMC2715816
- DOI: 10.1128/MCB.00185-09
Cyclin-dependent kinase inhibits reinitiation of a normal S-phase program during G2 in fission yeast
Abstract
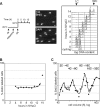
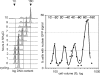
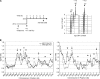
To achieve faithful replication of the genome once in each cell cycle, reinitiation of S phase is prevented in G(2) and origins are restricted from refiring within S phase. We have investigated the block to rereplication during G(2) in fission yeast. The DNA synthesis that occurs when G(2)/M cyclin-dependent kinase (CDK) activity is depleted has been assumed to be repeated rounds of S phase without mitosis, but this has not been demonstrated to be the case. We show here that on G(2)/M CDK depletion in G(2), repeated S phases are induced, which are correlated with normal G(1)/S transcription and attainment of doublings in cell size. Mostly normal mitotic S-phase origins are utilized, although at different efficiencies, and replication is essentially equal across the genome. We conclude that CDK inhibits reinitiation of S phase during G(2), and if G(2)/M CDK is depleted, replication results from induction of a largely normal S-phase program with only small differences in origin usage and efficiency.
Figures


References
-
- Arias, E. E., and J. C. Walter. 2007. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21497-518. - PubMed
-
- Ayté, J., C. Schweitzer, P. Zarzov, P. Nurse, and J. A. DeCaprio. 2001. Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat. Cell Biol. 31043-1050. - PubMed
-
- Bähler, J. 2005. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 3969-94. - PubMed
-
- Bähler, J., J.-Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14943-951. - PubMed
-
- Bates, S., K. M. Ryan, A. C. Phillips, and K. H. Vousden. 1998. Cell cycle arrest and DNA endoreduplication following p21Waf1/Cip1 expression. Oncogene 171691-1703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
