Sab, a novel autotransporter of locus of enterocyte effacement-negative shiga-toxigenic Escherichia coli O113:H21, contributes to adherence and biofilm formation
- PMID: 19487483
- PMCID: PMC2715694
- DOI: 10.1128/IAI.00031-09
Sab, a novel autotransporter of locus of enterocyte effacement-negative shiga-toxigenic Escherichia coli O113:H21, contributes to adherence and biofilm formation
Abstract
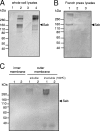
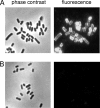
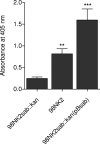
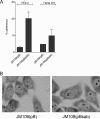
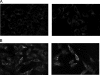
Shiga-toxigenic Escherichia coli (STEC) strains cause serious gastrointestinal disease, which can lead to potentially life-threatening systemic complications such as hemolytic-uremic syndrome. Although the production of Shiga toxin has been considered to be the main virulence trait of STEC for many years, the capacity to colonize the host intestinal epithelium is a crucial step in pathogenesis. In this study, we have characterized a novel megaplasmid-encoded outer membrane protein in locus of enterocyte effacement (LEE)-negative O113:H21 STEC strain 98NK2, termed Sab (for STEC autotransporter [AT] contributing to biofilm formation). The 4,296-bp sab gene encodes a 1,431-amino-acid protein with the features of members of the AT protein family. When expressed in E. coli JM109, Sab contributed to the diffuse adherence to human epithelial (HEp-2) cells and promoted biofilm formation on polystyrene surfaces. A 98NK2 sab deletion mutant was also defective in biofilm formation relative to its otherwise isogenic wild-type parent, and this was complemented by transformation with a sab-carrying plasmid. Interestingly, an unrelated O113:H21 STEC isolate that had a naturally occurring deletion in sab was similarly defective in biofilm formation. PCR analysis indicated that sab is present in LEE-negative STEC strains belonging to serotypes/groups O113:H21, O23, and O82:H8. These findings raise the possibility that Sab may contribute to colonization in a subset of LEE-negative STEC strains.
Figures
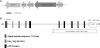

Similar articles
-
Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans.Infect Immun. 2001 Nov;69(11):6999-7009. doi: 10.1128/IAI.69.11.6999-7009.2001. Infect Immun. 2001. PMID: 11598075 Free PMC article.
-
Low prevalence of STEC autotransporter contributing to biofilm formation (Sab) in verocytotoxin-producing Escherichia coli isolates of humans and raw meats.Eur J Clin Microbiol Infect Dis. 2012 Jul;31(7):1463-5. doi: 10.1007/s10096-011-1464-y. Epub 2011 Nov 6. Eur J Clin Microbiol Infect Dis. 2012. PMID: 22057420
-
Enhanced CXC chemokine responses of human colonic epithelial cells to locus of enterocyte effacement-negative shiga-toxigenic Escherichia coli.Infect Immun. 2003 Oct;71(10):5623-32. doi: 10.1128/IAI.71.10.5623-5632.2003. Infect Immun. 2003. PMID: 14500482 Free PMC article.
-
Toxins of Locus of Enterocyte Effacement-Negative Shiga Toxin-Producing Escherichia coli.Toxins (Basel). 2018 Jun 14;10(6):241. doi: 10.3390/toxins10060241. Toxins (Basel). 2018. PMID: 29903982 Free PMC article. Review.
-
Detection of Shiga Toxin-Producing Escherichia coli from Nonhuman Sources and Strain Typing.Microbiol Spectr. 2014 Jun;2(3). doi: 10.1128/microbiolspec.EHEC-0001-2013. Microbiol Spectr. 2014. PMID: 26103970 Review.
Cited by
-
Prevalence and Implications of Shiga Toxin-Producing E. coli in Farm and Wild Ruminants.Pathogens. 2022 Nov 11;11(11):1332. doi: 10.3390/pathogens11111332. Pathogens. 2022. PMID: 36422584 Free PMC article. Review.
-
Characterization of Shiga toxin-producing Escherichia coli O130:H11 and O178:H19 isolated from dairy cows.Front Cell Infect Microbiol. 2013 Mar 8;3:9. doi: 10.3389/fcimb.2013.00009. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 23483233 Free PMC article.
-
SARS-CoV-2 and Emerging Foodborne Pathogens: Intriguing Commonalities and Obvious Differences.Pathogens. 2022 Jul 27;11(8):837. doi: 10.3390/pathogens11080837. Pathogens. 2022. PMID: 36014958 Free PMC article. Review.
-
Virulence Profiling and Molecular Typing of Shiga Toxin-Producing E. coli (STEC) from Human Sources in Brazil.Microorganisms. 2020 Jan 25;8(2):171. doi: 10.3390/microorganisms8020171. Microorganisms. 2020. PMID: 31991731 Free PMC article.
-
Genetic diversity and virulence potential of shiga toxin-producing Escherichia coli O113:H21 strains isolated from clinical, environmental, and food sources.Appl Environ Microbiol. 2014 Aug;80(15):4757-63. doi: 10.1128/AEM.01182-14. Epub 2014 May 23. Appl Environ Microbiol. 2014. PMID: 24858089 Free PMC article.
References
-
- Ackermann, N., M. Tiller, G. Anding, A. Roggenkamp, and J. Heesemann. 2008. Contribution of trimeric autotransporter C-terminal domains of oligomeric coiled-coil adhesin (Oca) family members YadA, UspA1, EibA, and Hia to translocation of the YadA passenger domain and virulence of Yersinia enterocolitica. J. Bacteriol. 1905031-5043. - PMC - PubMed
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. - PubMed
-
- Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24767-778. - PubMed
-
- Caron, E., V. F. Crepin, N. Simpson, S. Knutton, J. Garmendia, and G. Frankel. 2006. Subversion of actin dynamics by EPEC and EHEC. Curr. Opin. Microbiol. 940-45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

