Integration of microRNA miR-122 in hepatic circadian gene expression
- PMID: 19487572
- PMCID: PMC2701584
- DOI: 10.1101/gad.1781009
Integration of microRNA miR-122 in hepatic circadian gene expression
Abstract
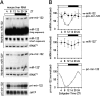
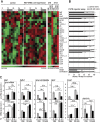
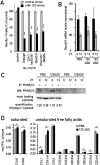
In liver, most metabolic pathways are under circadian control, and hundreds of protein-encoding genes are thus transcribed in a cyclic fashion. Here we show that rhythmic transcription extends to the locus specifying miR-122, a highly abundant, hepatocyte-specific microRNA. Genetic loss-of-function and gain-of-function experiments have identified the orphan nuclear receptor REV-ERBalpha as the major circadian regulator of mir-122 transcription. Although due to its long half-life mature miR-122 accumulates at nearly constant rates throughout the day, this miRNA is tightly associated with control mechanisms governing circadian gene expression. Thus, the knockdown of miR-122 expression via an antisense oligonucleotide (ASO) strategy resulted in the up- and down-regulation of hundreds of mRNAs, of which a disproportionately high fraction accumulates in a circadian fashion. miR-122 has previously been linked to the regulation of cholesterol and lipid metabolism. The transcripts associated with these pathways indeed show the strongest time point-specific changes upon miR-122 depletion. The identification of Pparbeta/delta and the peroxisome proliferator-activated receptor alpha (PPARalpha) coactivator Smarcd1/Baf60a as novel miR-122 targets suggests an involvement of the circadian metabolic regulators of the PPAR family in miR-122-mediated metabolic control.
Figures




Comment in
-
Liver "tick tock".Hepatology. 2009 Oct;50(4):1310-1. doi: 10.1002/hep.23266. Hepatology. 2009. PMID: 19790199 No abstract available.
References
-
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4:76–84. - PubMed
-
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. - PubMed
-
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. - PubMed
-
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
