The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo
- PMID: 19487573
- PMCID: PMC2701585
- DOI: 10.1101/gad.1777409
The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo
Abstract
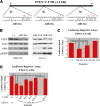
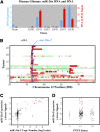
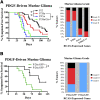
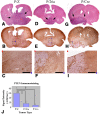
Activated oncogenic signaling is central to the development of nearly all forms of cancer, including the most common class of primary brain tumor, glioma. Research over the last two decades has revealed the particular importance of the Akt pathway, and its molecular antagonist PTEN (phosphatase and tensin homolog), in the process of gliomagenesis. Recent studies have also demonstrated that microRNAs (miRNAs) may be responsible for the modulation of cancer-implicated genes in tumors. Here we report the identification miR-26a as a direct regulator of PTEN expression. We also show that miR-26a is frequently amplified at the DNA level in human glioma, most often in association with monoallelic PTEN loss. Finally, we demonstrate that miR-26a-mediated PTEN repression in a murine glioma model both enhances de novo tumor formation and precludes loss of heterozygosity and the PTEN locus. Our results document a new epigenetic mechanism for PTEN regulation in glioma and further highlight dysregulation of Akt signaling as crucial to the development of these tumors.
Figures


References
-
- Baehring JM. An update on oligodendroglial neoplasms. Curr Opin Neurol. 2005;18:639–644. - PubMed
-
- Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta Neuropathol. 2003;106:479–485. - PubMed
-
- Becher OJ, Hambardzumyan D, Fomchenko EI, Momota H, Mainwaring L, Bleau AM, Katz AM, Edgar M, Kenney AM, Cordon-Cardo C, et al. Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer Res. 2008;68:2241–2249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
