Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta
- PMID: 19487673
- PMCID: PMC2701015
- DOI: 10.1073/pnas.0902175106
Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta
Abstract
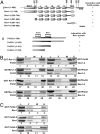
Yeast DNA polymerase (Pol) delta, essential for DNA replication, is comprised of 3 subunits, Pol3, Pol31, and Pol32. Of these, the catalytic subunit Pol3 and the second subunit Pol31 are essential, whereas the Pol32 subunit is not essential for DNA replication. Although Pol32 is an integral component of Pol delta, it is also required for translesion synthesis (TLS) by Pol zeta. To begin to decipher the bases of Pol32 involvement in Pol zeta-mediated TLS, here we examine whether Pol32 physically interacts with Pol zeta or its associated proteins and provide evidence for the physical interaction of Pol32 with Rev1. Rev1 plays an indispensable structural role in Pol zeta-mediated TLS and it binds the Rev3 catalytic subunit of Pol zeta. Here, we show that although Pol32 does not directly bind Pol zeta, Pol32 can bind the Rev1-Pol zeta complex through its interaction with Rev1. We find that Pol32 binding has no stimulatory effect on DNA synthesis either by Rev1 in the Rev1-Pol32 complex or by Pol zeta in the Pol zeta-Rev1-Pol32 complex, irrespective of whether proliferating cell nuclear antigen has been loaded onto DNA or not. We discuss evidence for the biological significance of Rev1 binding to Pol32 for Pol zeta function in TLS and suggest a structural role for Rev1 in modulating the binding of Pol zeta with Pol32 in Pol delta stalled at a lesion site.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differential regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae.DNA Repair (Amst). 2014 Dec;24:138-149. doi: 10.1016/j.dnarep.2014.04.013. Epub 2014 May 10. DNA Repair (Amst). 2014. PMID: 24819597 Free PMC article.
-
A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis.Nucleic Acids Res. 2012 Dec;40(22):11618-26. doi: 10.1093/nar/gks948. Epub 2012 Oct 12. Nucleic Acids Res. 2012. PMID: 23066099 Free PMC article.
-
The translesion DNA polymerases Pol ζ and Rev1 are activated independently of PCNA ubiquitination upon UV radiation in mutants of DNA polymerase δ.PLoS Genet. 2017 Dec 27;13(12):e1007119. doi: 10.1371/journal.pgen.1007119. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29281621 Free PMC article.
-
Eukaryotic DNA polymerase ζ.DNA Repair (Amst). 2015 May;29:47-55. doi: 10.1016/j.dnarep.2015.02.012. Epub 2015 Feb 19. DNA Repair (Amst). 2015. PMID: 25737057 Free PMC article. Review.
-
Roles of mutagenic translesion synthesis in mammalian genome stability, health and disease.DNA Repair (Amst). 2015 May;29:56-64. doi: 10.1016/j.dnarep.2015.01.001. Epub 2015 Jan 21. DNA Repair (Amst). 2015. PMID: 25655219 Review.
Cited by
-
Sml1 Inhibits the DNA Repair Activity of Rev1 in Saccharomyces cerevisiae during Oxidative Stress.Appl Environ Microbiol. 2020 Mar 18;86(7):e02838-19. doi: 10.1128/AEM.02838-19. Print 2020 Mar 18. Appl Environ Microbiol. 2020. PMID: 32005731 Free PMC article.
-
DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ.J Biol Chem. 2012 May 18;287(21):17281-17287. doi: 10.1074/jbc.M112.351122. Epub 2012 Mar 30. J Biol Chem. 2012. PMID: 22465957 Free PMC article.
-
Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila.PLoS Genet. 2012;8(4):e1002659. doi: 10.1371/journal.pgen.1002659. Epub 2012 Apr 19. PLoS Genet. 2012. PMID: 22532806 Free PMC article.
-
A sophisticated mechanism governs Pol ζ activity in response to replication stress.Nat Commun. 2024 Aug 31;15(1):7562. doi: 10.1038/s41467-024-52112-z. Nat Commun. 2024. PMID: 39215012 Free PMC article.
-
The importance of an interaction network for proper DNA polymerase ζ heterotetramer activity.Curr Genet. 2018 Jun;64(3):575-580. doi: 10.1007/s00294-017-0789-1. Epub 2017 Nov 30. Curr Genet. 2018. PMID: 29189894 Free PMC article. Review.
References
-
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. - PubMed
-
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science. 1999;283:1001–1004. - PubMed
-
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

