Determining DNA sequence specificity of natural and artificial transcription factors by cognate site identifier analysis
- PMID: 19488729
- PMCID: PMC5122672
- DOI: 10.1007/978-1-59745-483-4_41
Determining DNA sequence specificity of natural and artificial transcription factors by cognate site identifier analysis
Abstract
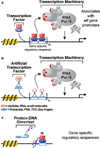
Artificial transcription factors (ATFs) are designed to mimic natural transcription factors in the control of gene expression and are comprised of domains for DNA binding and gene regulation. ATF domains are modular, interchangeable, and can be composed of protein-based or nonpeptidic moieties, yielding DNA-interacting regulatory molecules that can either activate or inhibit transcription. Sequence-specific targeting is a key determinant in ATF activity, and DNA-binding domains such as natural zinc fingers and synthetic polyamides have emerged as useful DNA targeting molecules. Defining the comprehensive DNA binding specificity of these targeting molecules for accurate manipulations of the genome can be achieved using cognate site identifier DNA microarrays to explore the entire sequence space of binding sites. Design of ATFs that regulate gene expression with temporal control will generate important molecular tools to probe cell- and tissue-specific gene regulation and to function as potential therapeutic agents.
Figures




References
-
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. - PubMed
-
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

