Epigenetic reprogramming by somatic cell nuclear transfer in primates
- PMID: 19489081
- PMCID: PMC2755529
- DOI: 10.1002/stem.60
Epigenetic reprogramming by somatic cell nuclear transfer in primates
Abstract
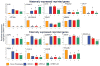
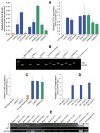
We recently demonstrated that somatic cells from adult primates could be reprogrammed into a pluripotent state by somatic cell nuclear transfer. However, the low efficiency with donor cells from one monkey necessitated the need for large oocyte numbers. Here, we demonstrate nearly threefold higher blastocyst development and embryonic stem (ES) cell derivation rates with different nuclear donor cells. Two ES cell lines were isolated using adult female rhesus macaque skin fibroblasts as nuclear donors and oocytes retrieved from one female, following a single controlled ovarian stimulation. In addition to routine pluripotency tests involving in vitro and in vivo differentiation into various somatic cell types, primate ES cells derived from reprogrammed somatic cells were also capable of contributing to cells expressing markers of germ cells. Moreover, imprinted gene expression, methylation, telomere length, and X-inactivation analyses were consistent with accurate and extensive epigenetic reprogramming of somatic cells by oocyte-specific factors.
Conflict of interest statement
Figures


References
-
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
-
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. - PubMed
-
- Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. - PubMed
-
- Byrne JA, Pedersen DA, Clepper LL, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

