A deletion and a duplication in distal 22q11.2 deletion syndrome region. Clinical implications and review
- PMID: 19490635
- PMCID: PMC2700091
- DOI: 10.1186/1471-2350-10-48
A deletion and a duplication in distal 22q11.2 deletion syndrome region. Clinical implications and review
Abstract
Background: Individuals affected with DiGeorge and Velocardiofacial syndromes present with both phenotypic diversity and variable expressivity. The most frequent clinical features include conotruncal congenital heart defects, velopharyngeal insufficiency, hypocalcemia and a characteristic craniofacial dysmorphism. The etiology in most patients is a 3 Mb recurrent deletion in region 22q11.2. However, cases of infrequent deletions and duplications with different sizes and locations have also been reported, generally with a milder, slightly different phenotype for duplications but with no clear genotype-phenotype correlation to date.
Methods: We present a 7 month-old male patient with surgically corrected ASD and multiple VSDs, and dysmorphic facial features not clearly suggestive of 22q11.2 deletion syndrome, and a newborn male infant with cleft lip and palate and upslanting palpebral fissures. Karyotype, FISH, MLPA, microsatellite markers segregation studies and SNP genotyping by array-CGH were performed in both patients and parents.
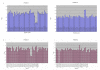
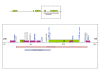
Results: Karyotype and FISH with probe N25 were normal for both patients. MLPA analysis detected a partial de novo 1.1 Mb deletion in one patient and a novel partial familial 0.4 Mb duplication in the other. Both of these alterations were located at a distal position within the commonly deleted region in 22q11.2. These rearrangements were confirmed and accurately characterized by microsatellite marker segregation studies and SNP array genotyping.
Conclusion: The phenotypic diversity found for deletions and duplications supports a lack of genotype-phenotype correlation in the vicinity of the LCRC-LCRD interval of the 22q11.2 chromosomal region, whereas the high presence of duplications in normal individuals supports their role as polymorphisms. We suggest that any hypothetical correlation between the clinical phenotype and the size and location of these alterations may be masked by other genetic and/or epigenetic modifying factors.
Figures


References
-
- Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer , Oechsler H, Belohradsky B, Prieur M, Aurias A, Raymond FL, Clayton-Smith J, Hatchwell E, McKeown C, Beemer FA, Dallapiccola B, Novelli G, Hurst JA, Ignatius J, Green AJ, Winter RM, Brueton L, Brondum-Nielsen K, Stewart F, Van Essen T, Patton M, Paterson J, Scambler PJ. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. - DOI - PMC - PubMed
-
- Carlson C, Sirotkin H, Pandita R, Goldberg R, McKie J, Wadey R, Patanjali SR, Weissman SM, Anyane-Yeboa K, Warburton D, Scambler P, Shprintzen R, Kucherlapati R, Morrow BE. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am J Hum Genet. 1997;61:620–629. doi: 10.1086/515508. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

