At loose ends: resecting a double-strand break
- PMID: 19490890
- PMCID: PMC3977653
- DOI: 10.1016/j.cell.2009.05.007
At loose ends: resecting a double-strand break
Abstract
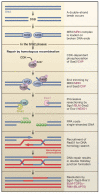
Double-strand break (DSB) repair is critical for maintaining genomic integrity and requires the processing of the 5' DSB ends. Recent studies have shed light on the mechanism and regulation of DNA end processing during DSB repair by homologous recombination.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

