TREAT Asia Quality Assessment Scheme (TAQAS) to standardize the outcome of HIV genotypic resistance testing in a group of Asian laboratories
- PMID: 19490972
- PMCID: PMC2863146
- DOI: 10.1016/j.jviromet.2009.03.016
TREAT Asia Quality Assessment Scheme (TAQAS) to standardize the outcome of HIV genotypic resistance testing in a group of Asian laboratories
Abstract
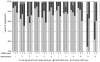
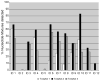
The TREAT Asia (Therapeutics, Research, Education, and AIDS Training in Asia) Network is building capacity for Human Immunodeficiency Virus Type-1 (HIV-1) drug resistance testing in the region. The objective of the TREAT Asia Quality Assessment Scheme - designated TAQAS - is to standardize HIV-1 genotypic resistance testing (HIV genotyping) among laboratories to permit rigorous comparison of results from different clinics and testing centres. TAQAS has evaluated three panels of HIV-1-positive plasma from clinical material or low-passage, culture supernatant for up to 10 Asian laboratories. Laboratory participants used their standard protocols to perform HIV genotyping. Assessment was in comparison to a target genotype derived from all participants and the reference laboratory's result. Agreement between most participants at the edited nucleotide sequence level was high (>98%). Most participants performed to the reference laboratory standard in detection of drug resistance mutations (DRMs). However, there was variation in the detection of nucleotide mixtures (0-83%) and a significant correlation with the detection of DRMs (p<0.01). Interpretation of antiretroviral resistance showed approximately 70% agreement among participants when different interpretation systems were used but >90% agreement with a common interpretation system, within the Stanford University Drug Resistance Database. Using the principles of external quality assessment and a reference laboratory, TAQAS has demonstrated high quality HIV genotyping results from Asian laboratories.
Figures

References
-
- Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organization’s global strategy for prevention and assessment of HIV drug resistance. Antiviral Ther. 2008;13:1–13. - PubMed
-
- de Luca A, Antinori A, di Giambenedetto S, Cingolani A, Perno CF, Cauda R. Interpretation systems for genotypic drug resistance of HIV-1. Scand J Infect Dis Suppl. 2003;106:29–34. - PubMed
-
- Demeter LM, D’Aquila R, Weislow O, Lorenzo E, Erice A, Fitzgibbon J, Shafer R, Richman D, Howard TM, Zhao Y, Fisher E, Huang D, Mayers D, Sylvester S, Arens M, Sannerud K, Rasheed S, Johnson V, Kuritzkes D, Reichelderfer P, Japour A ACTG Sequencing Working Group. AIDS Clinical Trials Group. Interlaboratory concordance of DNA sequence analysis to detect reverse transcriptase mutations in HIV-1 proviral DNA. J Virol Methods. 1998;75:93–104. - PubMed
-
- Descamps D, Delaugerre C, Masquelier B, Ruffault A, Marcelin AG, Izopet J, Chaix ML, Calvez V, Brun-Vezinet F, Costagliola D The ANTS Resistance Group. Repeated HIV-1 resistance genotyping external quality assessments improve virology laboratory performance. J Med Virol. 2006;78:159–160. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

