Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA
- PMID: 19491098
- PMCID: PMC2742811
- DOI: 10.1074/jbc.M109.020024
Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA
Abstract
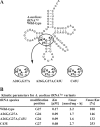
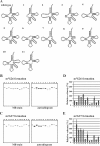
Transfer RNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes N2,N2-dimethylguanine formation at position 26 (m(2)(2)G26) in tRNA. In the reaction, N2-guanine at position 26 (m(2)G26) is generated as an intermediate. The trm1 genes are found only in archaea and eukaryotes, although it has been reported that Aquifex aeolicus, a hyper-thermophilic eubacterium, has a putative trm1 gene. To confirm whether A. aeolicus Trm1 has tRNA methyltransferase activity, we purified recombinant Trm1 protein. In vitro methyl transfer assay revealed that the protein has a strong tRNA methyltransferase activity. We confirmed that this gene product is expressed in living A. aeolicus cells and that the enzymatic activity exists in cell extract. By preparing 22 tRNA transcripts and testing their methyl group acceptance activities, it was demonstrated that this Trm1 protein has a novel tRNA specificity. Mass spectrometry analysis revealed that it catalyzes methyl transfers not only to G26 but also to G27 in substrate tRNA. Furthermore, it was confirmed that native tRNA(Cys) has an m(2)(2)G26m(2)G27 or m(2)(2)G26m(2)(2)G27 sequence, demonstrating that these modifications occur in living cells. Kinetic studies reveal that the m2G26 formation is faster than the m(2)G27 formation and that disruption of the G27-C43 base pair accelerates velocity of the G27 modification. Moreover, we prepared an additional 22 mutant tRNA transcripts and clarified that the recognition sites exist in the T-arm structure. This long distance recognition results in multisite recognition by the enzyme.
Figures






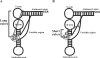
Similar articles
-
Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure.J Biol Chem. 2011 Oct 7;286(40):35236-46. doi: 10.1074/jbc.M111.253641. Epub 2011 Aug 15. J Biol Chem. 2011. PMID: 21844194 Free PMC article.
-
Aquifex aeolicus Trm1[tRNA (m2(2)G26) methyltransferase] has a novel recognition mechanism of the substrate RNA.Nucleic Acids Symp Ser (Oxf). 2005;(49):303-4. doi: 10.1093/nass/49.1.303. Nucleic Acids Symp Ser (Oxf). 2005. PMID: 17150754
-
Identification of Aquifex aeolicus tRNA (m2(2G26) methyltransferase gene.Nucleic Acids Res Suppl. 2002;(2):229-30. Nucleic Acids Res Suppl. 2002. PMID: 12903189
-
Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: importance of the tRNA architecture.Biochimie. 1995;77(1-2):54-61. doi: 10.1016/0300-9084(96)88104-1. Biochimie. 1995. PMID: 7599276 Review.
-
7-Methylguanosine Modifications in Transfer RNA (tRNA).Int J Mol Sci. 2018 Dec 17;19(12):4080. doi: 10.3390/ijms19124080. Int J Mol Sci. 2018. PMID: 30562954 Free PMC article. Review.
Cited by
-
Diversity in mechanism and function of tRNA methyltransferases.RNA Biol. 2015;12(4):398-411. doi: 10.1080/15476286.2015.1008358. RNA Biol. 2015. PMID: 25626150 Free PMC article. Review.
-
Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.Nucleic Acids Res. 2012 Jun;40(11):5149-61. doi: 10.1093/nar/gks163. Epub 2012 Feb 22. Nucleic Acids Res. 2012. PMID: 22362751 Free PMC article.
-
Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase.Nucleic Acids Res. 2010 Oct;38(18):6206-18. doi: 10.1093/nar/gkq381. Epub 2010 May 18. Nucleic Acids Res. 2010. PMID: 20483913 Free PMC article.
-
Methylated nucleosides in tRNA and tRNA methyltransferases.Front Genet. 2014 May 23;5:144. doi: 10.3389/fgene.2014.00144. eCollection 2014. Front Genet. 2014. PMID: 24904644 Free PMC article. Review.
-
Stereochemical mechanisms of tRNA methyltransferases.FEBS Lett. 2010 Jan 21;584(2):278-86. doi: 10.1016/j.febslet.2009.11.075. FEBS Lett. 2010. PMID: 19944101 Free PMC article. Review.
References
-
- Garcia G. R., Goodenough-Lashhua D. M. (1998) in Modification and Editing of RNA (Grosjean H., Benne R. eds) pp. 555–560, American Society for Microbiology,Washington, DC
-
- Constantinesco F., Motorin Y., Grosjean H. (1999) J. Mol. Biol. 291, 375–392 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

