Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression
- PMID: 19491106
- PMCID: PMC2742850
- DOI: 10.1074/jbc.M109.007336
Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression
Abstract
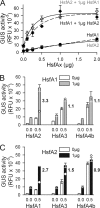
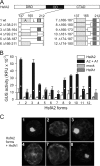
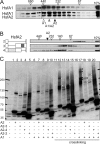
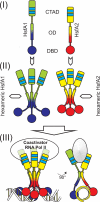
In plants, a family of more than 20 heat stress transcription factors (Hsf) controls the expression of heat stress (hs) genes. There is increasing evidence for the functional diversification between individual members of the Hsf family fulfilling distinct roles in response to various environmental stress conditions and developmental signals. In response to hs, accumulation of both heat stress proteins (Hsp) and Hsfs is induced. In tomato, the physical interaction between the constitutively expressed HsfA1 and the hs-inducible HsfA2 results in synergistic transcriptional activation (superactivation) of hs gene expression. Here, we show that the interaction is strikingly specific and not observed with other class A Hsfs. Hetero-oligomerization of the two-component Hsfs is preferred to homo-oligomerization, and each Hsf in the HsfA1/HsfA2 hetero-oligomeric complex has its characteristic contribution to its function as superactivator. Distinct regions of the oligomerization domain are responsible for specific homo- and hetero-oligomeric interactions leading to the formation of hexameric complexes. The results are summarized in a model of assembly and function of HsfA1/A2 superactivator complexes in hs gene regulation.
Figures



References
-
- Morimoto R. I. (1998) Genes Dev. 12, 3788–3796 - PubMed
-
- Baniwal S. K., Bharti K., Chan K. Y., Fauth M., Ganguli A., Kotak S., Mishra S. K., Nover L., Port M., Scharf K. D., Tripp J., Weber C., Zielinski D., von Koskull-Döring P. (2004) J. Biosci. 29, 471–487 - PubMed
-
- von Koskull-Döring P., Scharf K. D., Nover L. (2007) Trends Plant Sci. 12, 452–457 - PubMed
-
- Akerfelt M., Trouillet D., Mezger V., Sistonen L. (2007) Ann. N.Y. Acad. Sci. 1113, 15–27 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

