Helix straightening as an activation mechanism in the gelsolin superfamily of actin regulatory proteins
- PMID: 19491107
- PMCID: PMC2755850
- DOI: 10.1074/jbc.M109.019760
Helix straightening as an activation mechanism in the gelsolin superfamily of actin regulatory proteins
Abstract
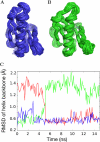
Villin and gelsolin consist of six homologous domains of the gelsolin/cofilin fold (V1-V6 and G1-G6, respectively). Villin differs from gelsolin in possessing at its C terminus an unrelated seventh domain, the villin headpiece. Here, we present the crystal structure of villin domain V6 in an environment in which intact villin would be inactive, in the absence of bound Ca(2+) or phosphorylation. The structure of V6 more closely resembles that of the activated form of G6, which contains one bound Ca(2+), rather than that of the calcium ion-free form of G6 within intact inactive gelsolin. Strikingly apparent is that the long helix in V6 is straight, as found in the activated form of G6, as opposed to the kinked version in inactive gelsolin. Molecular dynamics calculations suggest that the preferable conformation for this helix in the isolated G6 domain is also straight in the absence of Ca(2+) and other gelsolin domains. However, the G6 helix bends in intact calcium ion-free gelsolin to allow interaction with G2 and G4. We suggest that a similar situation exists in villin. Within the intact protein, a bent V6 helix, when triggered by Ca(2+), straightens and helps push apart adjacent domains to expose actin-binding sites within the protein. The sixth domain in this superfamily of proteins serves as a keystone that locks together a compact ensemble of domains in an inactive state. Perturbing the keystone initiates reorganization of the structure to reveal previously buried actin-binding sites.
Figures

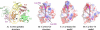


References
-
- dos Remedios C. G., Chhabra D., Kekic M., Dedova I. V., Tsubakihara M., Berry D. A., Nosworthy N. J. (2003) Physiol. Rev. 83, 433–473 - PubMed
-
- McGough A. M., Staiger C. J., Min J. K., Simonetti K. D. (2003) FEBS Lett. 552, 75–81 - PubMed
-
- Janmey P. A., Matsudaira P. T. (1988) J. Biol. Chem. 263, 16738–16743 - PubMed
-
- George S. P., Wang Y., Mathew S., Srinivasan K., Khurana S. (2007) J. Biol. Chem. 282, 26528–26541 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

