Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate
- PMID: 19491391
- PMCID: PMC2721792
- DOI: 10.1182/blood-2009-03-207944
Graded repression of PU.1/Sfpi1 gene transcription by GATA factors regulates hematopoietic cell fate
Abstract
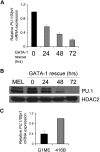
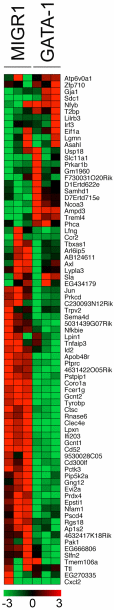
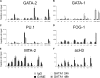
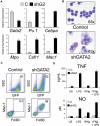
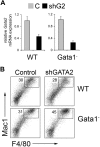
GATA-1 and PU.1 are essential hematopoietic transcription factors that control erythromegakaryocytic and myelolymphoid differentiation, respectively. These proteins antagonize each other through direct physical interaction to repress alternate lineage programs. We used immortalized Gata1(-) erythromegakaryocytic progenitor cells to study how PU.1/Sfpi1 expression is regulated by GATA-1 and GATA-2, a related factor that is normally expressed at earlier stages of hematopoiesis. Both GATA factors bind the PU.1/Sfpi1 gene at 2 highly conserved regions. In the absence of GATA-1, GATA-2 binding is associated with an undifferentiated state, intermediate level PU.1/Sfpi1 expression, and low-level expression of its downstream myeloid target genes. Restoration of GATA-1 function induces erythromegakaryocytic differentiation. Concomitantly, GATA-1 replaces GATA-2 at the PU.1/Sfpi1 locus and PU.1/Sfpi1 expression is extinguished. In contrast, when GATA-1 is not present, shRNA knockdown of GATA-2 increases PU.1/Sfpi1 expression by 3-fold and reprograms the cells to become macrophages. Our findings indicate that GATA factors act sequentially to regulate lineage determination during hematopoiesis, in part by exerting variable repressive effects at the PU.1/Sfpi1 locus.
Figures


References
-
- Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–738. - PubMed
-
- Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11:513–519. - PubMed
-
- Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26:6777–6794. - PubMed
-
- Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. - PubMed
-
- Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

