Combinatorial transcription factor regulation of the cyclic AMP-response element on the Pgc-1alpha promoter in white 3T3-L1 and brown HIB-1B preadipocytes
- PMID: 19491401
- PMCID: PMC2742839
- DOI: 10.1074/jbc.M109.021766
Combinatorial transcription factor regulation of the cyclic AMP-response element on the Pgc-1alpha promoter in white 3T3-L1 and brown HIB-1B preadipocytes
Abstract
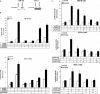
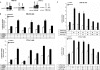
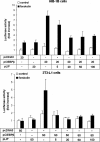
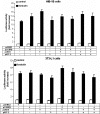
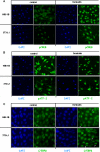
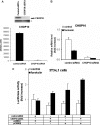
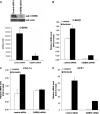
Cold stress in rodents increases the expression of UCP1 and PGC-1alpha in brown and white adipose tissue. We have previously reported that C/EBPbeta specifically binds to the CRE on the proximal Pgc-1alpha promoter and increases forskolin-sensitive Pgc-1alpha and Ucp1 expression in white 3T3-L1 preadipocytes. Here we show that in mice exposed to a cold environment for 24 h, Pgc-1alpha, Ucp1, and C/ebpbeta but not C/ebpalpha or C/ebpdelta expression were increased in BAT. Conversely, expression of the C/EBP dominant negative Chop10 was increased in WAT but not BAT during cold exposure. Reacclimatization of cold-exposed mice to a warm environment for 24 h completely reversed these changes in gene expression. In HIB-1B, brown preadipocytes, forskolin increased expression of Pgc-1alpha, Ucp1, and C/ebpbeta early in differentiation and inhibited Chop10 expression. Employing chromatin immunoprecipitation, we demonstrate that C/EBPbeta, CREB, ATF-2, and CHOP10 are bound to the Pgc-1alpha proximal CRE, but CHOP10 does not bind in HIB-1B cell lysates. Forskolin stimulation and C/EBPbeta overexpression in 3T3-L1 cells increased C/EBPbeta and CREB but displaced ATF-2 and CHOP10 binding to the Pgc-1alpha proximal CRE. Overexpression of ATF-2 and CHOP10 in 3T3-L1 cells decreased Pgc-1alpha transcription. Knockdown of Chop10 in 3T3-L1 cells using siRNA increased Pgc-1alpha transcription, whereas siRNA against C/ebpbeta in HIB-1B cells decreased Pgc-1alpha and Ucp1 expression. We conclude that the increased cAMP stimulation of Pgc-1alpha expression is regulated by the combinatorial effect of transcription factors acting at the CRE on the proximal Pgc-1alpha promoter.
Figures
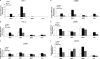


Similar articles
-
C/EBPbeta reprograms white 3T3-L1 preadipocytes to a Brown adipocyte pattern of gene expression.J Biol Chem. 2007 Aug 24;282(34):24660-9. doi: 10.1074/jbc.M703101200. Epub 2007 Jun 21. J Biol Chem. 2007. PMID: 17584738
-
Differentiation and characterization in primary culture of white adipose tissue brown adipocyte-like cells.Int J Obes (Lond). 2009 Jun;33(6):680-6. doi: 10.1038/ijo.2009.46. Epub 2009 Mar 10. Int J Obes (Lond). 2009. PMID: 19274054
-
Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1alpha.Mol Cell Biol. 2005 Feb;25(4):1272-82. doi: 10.1128/MCB.25.4.1272-1282.2005. Mol Cell Biol. 2005. PMID: 15684380 Free PMC article.
-
Transcriptional regulation of the uncoupling protein-1 gene.Biochimie. 2017 Mar;134:86-92. doi: 10.1016/j.biochi.2016.09.017. Epub 2016 Oct 5. Biochimie. 2017. PMID: 27693079 Review.
-
Turning WAT into BAT: a review on regulators controlling the browning of white adipocytes.Biosci Rep. 2013 Sep 6;33(5):e00065. doi: 10.1042/BSR20130046. Biosci Rep. 2013. PMID: 23895241 Free PMC article. Review.
Cited by
-
Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue.Diabetologia. 2010 Jun;53(6):1164-73. doi: 10.1007/s00125-010-1701-4. Epub 2010 Mar 18. Diabetologia. 2010. PMID: 20238096 Free PMC article.
-
Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) α and δ in brown adipocytes to match fatty acid oxidation with supply.J Biol Chem. 2012 Jul 20;287(30):25038-48. doi: 10.1074/jbc.M112.374041. Epub 2012 Jun 8. J Biol Chem. 2012. PMID: 22685301 Free PMC article.
-
Cold-induced changes in gene expression in brown adipose tissue, white adipose tissue and liver.PLoS One. 2013 Jul 22;8(7):e68933. doi: 10.1371/journal.pone.0068933. Print 2013. PLoS One. 2013. PMID: 23894377 Free PMC article.
-
Endoplasmic Reticulum Stress-Induced CHOP Inhibits PGC-1α and Causes Mitochondrial Dysfunction in Diabetic Embryopathy.Toxicol Sci. 2017 Aug 1;158(2):275-285. doi: 10.1093/toxsci/kfx096. Toxicol Sci. 2017. PMID: 28482072 Free PMC article.
-
Ovariectomy Interferes with Proteomes of Brown Adipose Tissue in Rats.Int J Med Sci. 2022 Mar 6;19(3):499-510. doi: 10.7150/ijms.66996. eCollection 2022. Int J Med Sci. 2022. PMID: 35370469 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

