Genetic Factors and Orofacial Clefting
- PMID: 19492008
- PMCID: PMC2598422
- DOI: 10.1053/j.sodo.2008.02.002
Genetic Factors and Orofacial Clefting
Abstract
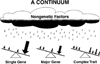
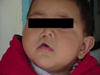
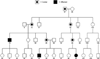
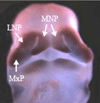
Cleft lip with or without cleft palate is the most common facial birth defect and it is caused by a complex interaction between genetic and environmental factors. The purpose of this review is to provide an overview of the spectrum of the genetic causes for cleft lip and cleft palate using both syndromic and nonsyndromic forms of clefting as examples. Although the gene identification process for orofacial clefting in humans is in the early stages, the pace is rapidly accelerating. Recently, several genes have been identified that have a combined role in up to 20% of all clefts. While this is a significant step forward, it is apparent that additional cleft causing genes have yet to be identified. Ongoing human genome-wide linkage studies have identified regions in the genome that likely contain genes that when mutated cause orofacial clefting, including a major gene on chromosome 9 that is positive in multiple racial groups. Currently, efforts are focused to identify which genes are mutated in these regions. In addition, parallel studies are also evaluating genes involved in environmental pathways. Furthermore, statistical geneticists are developing new methods to characterize both gene-gene and gene-environment interactions to build better models for pathogenesis of this common birth defect. The ultimate goal of these studies is to provide knowledge for more accurate risk counseling and the development of preventive therapies.
Figures
Similar articles
-
Orofacial Clefts: Genetics of Cleft Lip and Palate.Genes (Basel). 2023 Aug 9;14(8):1603. doi: 10.3390/genes14081603. Genes (Basel). 2023. PMID: 37628654 Free PMC article. Review.
-
Genome-wide analysis of copy-number variation in humans with cleft lip and/or cleft palate identifies COBLL1, RIC1, and ARHGEF38 as clefting genes.Am J Hum Genet. 2023 Jan 5;110(1):71-91. doi: 10.1016/j.ajhg.2022.11.012. Epub 2022 Dec 8. Am J Hum Genet. 2023. PMID: 36493769 Free PMC article.
-
Characteristics of Orofacial Clefting in Hawai'i.Hawaii J Health Soc Welf. 2019 Aug;78(8):258-261. Hawaii J Health Soc Welf. 2019. PMID: 31463475 Free PMC article.
-
IRF6 polymorphisms in Brazilian patients with non-syndromic cleft lip with or without palate.Braz J Otorhinolaryngol. 2020 Nov-Dec;86(6):696-702. doi: 10.1016/j.bjorl.2019.04.011. Epub 2019 Jun 8. Braz J Otorhinolaryngol. 2020. PMID: 31495697 Free PMC article.
-
The spectrum of orofacial clefting.Plast Reconstr Surg. 2005 Jun;115(7):101e-114e. doi: 10.1097/01.prs.0000164494.45986.91. Plast Reconstr Surg. 2005. PMID: 15923821 Review.
Cited by
-
The effects of oral clefts on hospital use throughout the lifespan.BMC Health Serv Res. 2012 Mar 9;12:58. doi: 10.1186/1472-6963-12-58. BMC Health Serv Res. 2012. PMID: 22405490 Free PMC article.
-
Identification of a novel heterozygous truncation mutation in exon 1 of ARHGAP29 in an Indian subject with nonsyndromic cleft lip with cleft palate.Eur J Dent. 2014 Oct;8(4):528-532. doi: 10.4103/1305-7456.143637. Eur J Dent. 2014. PMID: 25512736 Free PMC article.
-
The mechanism of TGF-β signaling during palate development.Oral Dis. 2011 Nov;17(8):733-44. doi: 10.1111/j.1601-0825.2011.01806.x. Epub 2011 Mar 13. Oral Dis. 2011. PMID: 21395922 Free PMC article. Review.
-
[Study of patients with cleft lip and palate with consanguineous parents].Braz J Otorhinolaryngol. 2011 Jan-Feb;77(1):19-23. doi: 10.1590/S1808-86942011000100004. Braz J Otorhinolaryngol. 2011. PMID: 21340184 Free PMC article.
-
A metabonomic approach to analyze the dexamethasone-induced cleft palate in mice.J Biomed Biotechnol. 2011;2011:509043. doi: 10.1155/2011/509043. Epub 2010 Aug 10. J Biomed Biotechnol. 2011. PMID: 20814536 Free PMC article.
References
-
- Cohen MM. Syndromes with cleft lip and cleft palate. Cleft Palate J. 1978;15:306–328. - PubMed
-
- Shprintzen RJ, Siegel VL, Amato J, et al. Anomalies associated with cleft lip, cleft palate, or both. American Journal of Medical Genetics. 1985;20:585–595. - PubMed
-
- Gorlin RJ. Syndromes of the head and neck. New York: Oxford University Press; 1990.
-
- Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. - PubMed
-
- Marazita ML, Mooney MP. Current concepts in the embryology and genetics of cleft lip and cleft palate. Clin Plast Surg. 2004;31:125–140. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
