Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis
- PMID: 19492050
- PMCID: PMC2685984
- DOI: 10.1371/journal.pone.0005635
Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis
Abstract
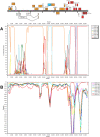
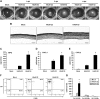
In 2005, a human adenovirus strain (formerly known as HAdV-D22/H8 but renamed here HAdV-D53) was isolated from an outbreak of epidemic keratoconjunctititis (EKC), a disease that is usually caused by HAdV-D8, -D19, or -D37, not HAdV-D22. To date, a complete change of tropism compared to the prototype has never been observed, although apparent recombinant strains of other viruses from species Human adenovirus D (HAdV-D) have been described. The complete genome of HAdV-D53 was sequenced to elucidate recombination events that lead to the emergence of a viable and highly virulent virus with a modified tropism. Bioinformatic and phylogenetic analyses of this genome demonstrate that this adenovirus is a recombinant of HAdV-D8 (including the fiber gene encoding the primary cellular receptor binding site), HAdV-D22, (the epsilon determinant of the hexon gene), HAdV-D37 (including the penton base gene encoding the secondary cellular receptor binding site), and at least one unknown or unsequenced HAdV-D strain. Bootscanning analysis of the complete genomic sequence of this novel adenovirus, which we have re-named HAdV-D53, indicated at least five recombination events between the aforementioned adenoviruses. Intrahexon recombination sites perfectly framed the epsilon neutralization determinant that was almost identical to the HAdV-D22 prototype. Additional bootscan analysis of all HAdV-D hexon genes revealed recombinations in identical locations in several other adenoviruses. In addition, HAdV-D53 but not HAdV-D22 induced corneal inflammation in a mouse model. Serological analysis confirmed previous results and demonstrated that HAdV-D53 has a neutralization profile representative of the epsilon determinant of its hexon (HAdV-D22) and the fiber (HAdV-D8) proteins. Our recombinant hexon sequence is almost identical to the hexon sequences of the HAdV-D strain causing EKC outbreaks in Japan, suggesting that HAdV-D53 is pandemic as an emerging EKC agent. This documents the first genomic, bioinformatic, and biological descriptions of the molecular evolution events engendering an emerging pathogenic adenovirus.
Conflict of interest statement
Figures




References
-
- Gordon YJ, Aoki K, Kinchington PR. Adenovirus keratoconjunctivitis. In: Pepose JS, Holland GN, Wilhelmus KR, editors. Ocular Infection and Immunity. St. Louis: Mosby; 1996. pp. 877–94.
-
- Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med. 1953;84:570–573. - PubMed
-
- Hilleman MR, Werner JH. Recovery of new agent from patients with acute respiratory illness. Proc Soc Exp Biol Med. 1954;851:183–188. - PubMed
-
- Dingle JH, Langmuir AD. Epidemiology of acute, respiratory disease in military recruits. Am Rev Respir Dis. 1968;976(Suppl):1–65. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

