Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion
- PMID: 19492051
- PMCID: PMC2685986
- DOI: 10.1371/journal.pone.0005780
Surface-associated plasminogen binding of Cryptococcus neoformans promotes extracellular matrix invasion
Abstract
Background: The fungal pathogen Cryptococcus neoformans is a leading cause of illness and death in persons with predisposing factors, including: malignancies, solid organ transplants, and corticosteroid use. C. neoformans is ubiquitous in the environment and enters into the lungs via inhalation, where it can disseminate through the bloodstream and penetrate the central nervous system (CNS), resulting in a difficult to treat and often-fatal infection of the brain, called meningoencephalitis. Plasminogen is a highly abundant protein found in the plasma component of blood and is necessary for the degradation of fibrin, collagen, and other structural components of tissues. This fibrinolytic system is utilized by cancer cells during metastasis and several pathogenic species of bacteria have been found to manipulate the host plasminogen system to facilitate invasion of tissues during infection by modifying the activation of this process through the binding of plasminogen at their surface.
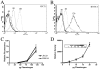
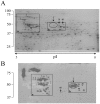
Methodology: The invasion of the brain and the central nervous system by penetration of the protective blood-brain barrier is a prerequisite to the establishment of meningoencephalitis by the opportunistic fungal pathogen C. neoformans. In this study, we examined the ability of C. neoformans to subvert the host plasminogen system to facilitate tissue barrier invasion. Through a combination of biochemical, cell biology, and proteomic approaches, we have shown that C. neoformans utilizes the host plasminogen system to cross tissue barriers, providing support for the hypothesis that plasminogen-binding may contribute to the invasion of the blood-brain barrier by penetration of the brain endothelial cells and underlying matrix. In addition, we have identified the cell wall-associated proteins that serve as plasminogen receptors and characterized both the plasminogen-binding and plasmin-activation potential for this significant human pathogen.
Conclusions: The results of this study provide evidence for the cooperative role of multiple virulence determinants in C. neoformans pathogenesis and suggest new avenues for the development of anti-infective agents in the prevention of fungal tissue invasion.
Conflict of interest statement
Figures
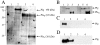
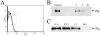



Similar articles
-
Cryptococcus neoformans Infection in the Central Nervous System: The Battle between Host and Pathogen.J Fungi (Basel). 2022 Oct 12;8(10):1069. doi: 10.3390/jof8101069. J Fungi (Basel). 2022. PMID: 36294634 Free PMC article. Review.
-
Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease.mBio. 2014 Jun 3;5(3):e01101-14. doi: 10.1128/mBio.01101-14. mBio. 2014. PMID: 24895304 Free PMC article.
-
Induction of brain microvascular endothelial cell urokinase expression by Cryptococcus neoformans facilitates blood-brain barrier invasion.PLoS One. 2012;7(11):e49402. doi: 10.1371/journal.pone.0049402. Epub 2012 Nov 8. PLoS One. 2012. PMID: 23145170 Free PMC article.
-
Blood-brain barrier invasion by Cryptococcus neoformans is enhanced by functional interactions with plasmin.Microbiology (Reading). 2012 Jan;158(Pt 1):240-258. doi: 10.1099/mic.0.051524-0. Epub 2011 Oct 13. Microbiology (Reading). 2012. PMID: 21998162 Free PMC article.
-
[Mechanism of Cryptococcus Meningoencephalitis].Med Mycol J. 2016;57(1):J27-32. doi: 10.3314/mmj.57.J27. Med Mycol J. 2016. PMID: 26936349 Review. Japanese.
Cited by
-
Bacillus anthracis interacts with plasmin(ogen) to evade C3b-dependent innate immunity.PLoS One. 2011 Mar 25;6(3):e18119. doi: 10.1371/journal.pone.0018119. PLoS One. 2011. PMID: 21464960 Free PMC article.
-
Multifunctional Fructose 1,6-Bisphosphate Aldolase as a Therapeutic Target.Front Mol Biosci. 2021 Aug 11;8:719678. doi: 10.3389/fmolb.2021.719678. eCollection 2021. Front Mol Biosci. 2021. PMID: 34458323 Free PMC article. Review.
-
The pathways and the mechanisms by which Cryptococcus enters the brain.Mycology. 2024 Feb 14;15(3):345-359. doi: 10.1080/21501203.2023.2295409. eCollection 2024. Mycology. 2024. PMID: 39247889 Free PMC article. Review.
-
Cryptococcus neoformans Infection in the Central Nervous System: The Battle between Host and Pathogen.J Fungi (Basel). 2022 Oct 12;8(10):1069. doi: 10.3390/jof8101069. J Fungi (Basel). 2022. PMID: 36294634 Free PMC article. Review.
-
The Response of Paracoccidioides lutzii to the Interaction with Human Neutrophils.J Fungi (Basel). 2023 Nov 7;9(11):1088. doi: 10.3390/jof9111088. J Fungi (Basel). 2023. PMID: 37998893 Free PMC article.
References
-
- Chen SH, Stins MF, Huang SH, Chen YH, Kwon-Chung KJ, et al. Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J Med Microbiol. 2003;52(Pt 11):961–970. - PubMed
-
- Chretien F, Lortholary O, Kansau I, Neuville S, Gray F, et al. Pathogenesis of cerebral Cryptococcus neoformans infection after fungemia. J Infect Dis. 2002;186(4):522–530. - PubMed
-
- Huang SH, Jong AY. Cellular mechanisms of microbial proteins contributing to invasion of the blood-brain barrier. Cell Microbiol. 2001;3(5):277–287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

