Structural and functional analysis of Sulfolobus solfataricus Y-family DNA polymerase Dpo4-catalyzed bypass of the malondialdehyde-deoxyguanosine adduct
- PMID: 19492857
- PMCID: PMC2717710
- DOI: 10.1021/bi9003588
Structural and functional analysis of Sulfolobus solfataricus Y-family DNA polymerase Dpo4-catalyzed bypass of the malondialdehyde-deoxyguanosine adduct
Abstract
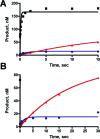
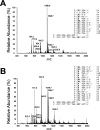
Oxidative stress can induce the formation of reactive electrophiles, such as DNA peroxidation products, e.g., base propenals, and lipid peroxidation products, e.g., malondialdehyde. Base propenals and malondialdehyde react with DNA to form adducts, including 3-(2'-deoxy-beta-D-erythro-pentofuranosyl)pyrimido[1,2-alpha]purin-10(3H)-one (M1dG). When paired opposite cytosine in duplex DNA at physiological pH, M1dG undergoes ring opening to form N2-(3-oxo-1-propenyl)-dG (N2-OPdG). Previous work has shown that M1dG is mutagenic in bacteria and mammalian cells and that its mutagenicity in Escherichia coli is dependent on induction of the SOS response, indicating a role for translesion DNA polymerases in the bypass of M1dG. To probe the mechanism by which translesion polymerases bypass M1dG, kinetic and structural studies were conducted with a model Y-family DNA polymerase, Dpo4 from Sulfolobus solfataricus. The level of steady-state incorporation of dNTPs opposite M1dG was reduced 260-2900-fold and exhibited a preference for dATP incorporation. Liquid chromatography-tandem mass spectrometry analysis of the full-length extension products revealed a spectrum of products arising principally by incorporation of dC or dA opposite M1dG followed by partial or full-length extension. A greater proportion of -1 deletions were observed when dT was positioned 5' of M1dG. Two crystal structures were determined, including a "type II" frameshift deletion complex and another complex with Dpo4 bound to a dC.M1dG pair located in the postinsertion context. Importantly, M1dG was in the ring-closed state in both structures, and in the structure with dC opposite M1dG, the dC residue moved out of the Dpo4 active site, into the minor groove. The results are consistent with the reported mutagenicity of M1dG and illustrate how the lesion may affect replication events.
Figures

Similar articles
-
Ring-opening of the γ-OH-PdG adduct promotes error-free bypass by the Sulfolobus solfataricus DNA polymerase Dpo4.Chem Res Toxicol. 2013 Sep 16;26(9):1348-60. doi: 10.1021/tx400200b. Epub 2013 Aug 16. Chem Res Toxicol. 2013. PMID: 23947567 Free PMC article.
-
Translesion DNA synthesis by human DNA polymerase eta on templates containing a pyrimidopurinone deoxyguanosine adduct, 3-(2'-deoxy-beta-d-erythro-pentofuranosyl)pyrimido-[1,2-a]purin-10(3H)-one.Biochemistry. 2009 Jan 20;48(2):471-80. doi: 10.1021/bi801591a. Biochemistry. 2009. PMID: 19108641 Free PMC article.
-
In vitro bypass of malondialdehyde-deoxyguanosine adducts: differential base selection during extension by the Klenow fragment of DNA polymerase I is the critical determinant of replication outcome.Biochemistry. 2004 Sep 21;43(37):11828-35. doi: 10.1021/bi049360f. Biochemistry. 2004. PMID: 15362868
-
A rescue act: Translesion DNA synthesis past N(2) -deoxyguanosine adducts.IUBMB Life. 2015 Jul;67(7):564-74. doi: 10.1002/iub.1403. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26173005 Review.
-
Recent insight into the kinetic mechanisms and conformational dynamics of Y-Family DNA polymerases.Biochemistry. 2014 May 6;53(17):2804-14. doi: 10.1021/bi5000405. Epub 2014 Apr 23. Biochemistry. 2014. PMID: 24716482 Free PMC article. Review.
Cited by
-
Escherichia coli Fpg glycosylase is nonrendundant and required for the rapid global repair of oxidized purine and pyrimidine damage in vivo.J Mol Biol. 2011 Jul 8;410(2):183-93. doi: 10.1016/j.jmb.2011.05.004. Epub 2011 May 13. J Mol Biol. 2011. PMID: 21601577 Free PMC article.
-
Translesion synthesis across 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2'-deoxyadenosine (1,N6-γ-HMHP-dA) adducts by human and archebacterial DNA polymerases.J Biol Chem. 2012 Nov 9;287(46):38800-11. doi: 10.1074/jbc.M112.396788. Epub 2012 Sep 13. J Biol Chem. 2012. PMID: 22977231 Free PMC article.
-
Computational Evaluation of Nucleotide Insertion Opposite Expanded and Widened DNA by the Translesion Synthesis Polymerase Dpo4.Molecules. 2016 Jun 23;21(7):822. doi: 10.3390/molecules21070822. Molecules. 2016. PMID: 27347908 Free PMC article.
-
Structural and kinetic insights into binding and incorporation of L-nucleotide analogs by a Y-family DNA polymerase.Nucleic Acids Res. 2014 Sep;42(15):9984-95. doi: 10.1093/nar/gku709. Epub 2014 Aug 7. Nucleic Acids Res. 2014. PMID: 25104018 Free PMC article.
-
In vitro bypass of the major malondialdehyde- and base propenal-derived DNA adduct by human Y-family DNA polymerases κ, ι, and Rev1.Biochemistry. 2010 Sep 28;49(38):8415-24. doi: 10.1021/bi1009024. Biochemistry. 2010. PMID: 20726503 Free PMC article.
References
-
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Shultz R. A., and Ellenberger T. (2006) DNA Repair and Mutagenesis, 2nd ed., ASM Press, Washington, DC.
-
- Basu A. K.; O'Hara S. M.; Valladier P.; Stone K.; Mols O.; Marnett L. J. (1988) Identification of adducts formed by reaction of guanine nucleosides with malondialdehyde and structurally related aldehydes. Chem. Res. Toxicol. 1, 53–59. - PubMed
-
- Seto H.; Okuda T.; Takesue T.; Ikemura T. (1983) Reaction of malonaldehyde with nucleic acid. I. Formation of fluorescent pyrimido[1,2a]purin-10(3H)-one nucleosides. Bull. Chem. Soc. Jpn. 56, 1799–1802.
-
- Chaudhary A. K.; Nokubo M.; Reddy G. R.; Yeola S. N.; Morrow J. D.; Blair I. A.; Marnett L. J. (1994) Detection of endogenous malondialdehyde-deoxyguanosine adducts in human liver. Science 265, 1580–1582. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

