Cross-study analysis of genomic data defines the ciliate multigenic epiplasmin family: strategies for functional analysis in Paramecium tetraurelia
- PMID: 19493334
- PMCID: PMC2709106
- DOI: 10.1186/1471-2148-9-125
Cross-study analysis of genomic data defines the ciliate multigenic epiplasmin family: strategies for functional analysis in Paramecium tetraurelia
Abstract
Background: The sub-membranous skeleton of the ciliate Paramecium, the epiplasm, is composed of hundreds of epiplasmic scales centered on basal bodies, and presents a complex set of proteins, epiplasmins, which belong to a multigenic family. The repeated duplications observed in the P. tetraurelia genome present an interesting model of the organization and evolution of a multigenic family within a single cell.
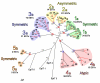
Results: To study this multigenic family, we used phylogenetic, structural, and analytical transcriptional approaches. The phylogenetic method defines 5 groups of epiplasmins in the multigenic family. A refined analysis by Hydrophobic Cluster Analysis (HCA) identifies structural characteristics of 51 epiplasmins, defining five separate groups, and three classes. Depending on the sequential arrangement of their structural domains, the epiplasmins are defined as symmetric, asymmetric or atypical. The EST data aid in this classification, in the identification of putative regulating sequences such as TATA or CAAT boxes. When specific RNAi experiments were conducted using sequences from either symmetric or asymmetric classes, phenotypes were drastic. Local effects show either disrupted or ill-shaped epiplasmic scales. In either case, this results in aborted cell division. Using structural features, we show that 4 epiplasmins are also present in another ciliate, Tetrahymena thermophila. Their affiliation with the distinctive structural groups of Paramecium epiplasmins demonstrates an interspecific multigenic family.
Conclusion: The epiplasmin multigenic family illustrates the history of genomic duplication in Paramecium. This study provides a framework which can guide functional analysis of epiplasmins, the major components of the membrane skeleton in ciliates. We show that this set of proteins handles an important developmental information in Paramecium since maintenance of epiplasm organization is crucial for cell morphogenesis.
Figures





References
-
- Keryer G, Adoutte A, Ng SF, Cohen J, Deloubresse NG, Rossignol M, Stelly N, Beisson J. Purification of the surface membrane-cytoskeleton complex (cortex) of Paramecium and identification of several of its protein constituents. European Journal of Protistology. 1990;25:209–225. - PubMed
-
- Iftode F, Cohen J, Ruiz F, Rueda AT, Chenshan L, Adoutte A, Beisson J. Development of surface pattern during division in Paramecium .1. Mapping of Duplication and Reorganization of cortical cytoskeletal structures in the wild-type. Development. 1989;105:191–211.
-
- Nahon P, Coffe G, Leguyader H, Darmanadendelorme J, Jeanmairewolf R, Clerot JC, Adoutte A. Identification of the epiplasmins, a new set of cortical proteins of the membrane cytoskeleton in Paramecium. Journal of Cell Science. 1993;104:975–990.
-
- Coffe G, LeCaer JP, Lima O, Adoutte A. Purification, in vitro reassembly, and preliminary sequence analysis of epiplasmins, the major constituent of the membrane skeleton of Paramecium. Cell Motility and the Cytoskeleton. 1996;34:137–151. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

