Protein intrinsic disorder and influenza virulence: the 1918 H1N1 and H5N1 viruses
- PMID: 19493338
- PMCID: PMC2701943
- DOI: 10.1186/1743-422X-6-69
Protein intrinsic disorder and influenza virulence: the 1918 H1N1 and H5N1 viruses
Abstract
Background: The 1918 H1N1 virus was a highly virulent strain that killed 20-50 million people. The cause of its virulence remains poorly understood.
Methods: Intrinsic disorder predictor PONDR VLXT was used to compare various influenza subtypes and strains. Three-dimensional models using data from X-ray crystallographic studies annotated with disorder prediction were used to characterize the proteins.
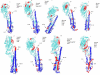
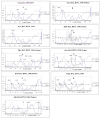
Results: The protein of interest is hemagglutin (HA), which is a surface glycoprotein that plays a vital role in viral entry. Distinct differences between HA proteins of the virulent and non-virulent strains are seen, especially in the region near residues 68-79 of the HA2. This region represents the tip of the stalk that is in contact with the receptor chain, HA1, and therefore likely to provide the greatest effect on the motions of the exposed portion of HA. Comparison of this region between virulent strains (1918 H1N1 and H5N1) and less virulent ones (H3N2 and 1930 H1N1) reveals that predicted disorder can be seen at this region among the more virulent strains and subtypes but is remarkably absent among the distinctly less virulent ones.
Conclusion: The motions created by disorder at crucial regions are likely to impair recognition by immunological molecules and increase the virulence of both the H5N1 and the 1918 H1N1 viruses. The results help explain many puzzling features of the H5N1 and the 1918 H1N1 viruses. Summarizing, HA (and especially its intrinsically disordered regions) can serve as a predictor of the influenza A virulence, even though there may be other proteins that contribute to or exacerbate the virulence.
Figures

Similar articles
-
A novel pathogenic mechanism of highly pathogenic avian influenza H5N1 viruses involves hemagglutinin mediated resistance to serum innate inhibitors.PLoS One. 2012;7(5):e36318. doi: 10.1371/journal.pone.0036318. Epub 2012 May 1. PLoS One. 2012. PMID: 22563489 Free PMC article.
-
Comparison of the pathology caused by H1N1, H5N1, and H3N2 influenza viruses.Arch Med Res. 2009 Nov;40(8):655-61. doi: 10.1016/j.arcmed.2009.10.001. Epub 2010 Jan 6. Arch Med Res. 2009. PMID: 20304252 Review.
-
Differential host determinants contribute to the pathogenesis of 2009 pandemic H1N1 and human H5N1 influenza A viruses in experimental mouse models.Am J Pathol. 2011 Jul;179(1):230-9. doi: 10.1016/j.ajpath.2011.03.041. Epub 2011 May 18. Am J Pathol. 2011. PMID: 21703405 Free PMC article.
-
[Virological impact of stalk region of neuraminidase in influenza A/Anhui/1/05 (H5N1) and A/Ohio/07/2009 (H1N1) viruses].Bing Du Xue Bao. 2014 May;30(3):238-45. Bing Du Xue Bao. 2014. PMID: 25118377 Chinese.
-
Innate immune responses to influenza A H5N1: friend or foe?Trends Immunol. 2009 Dec;30(12):574-84. doi: 10.1016/j.it.2009.09.004. Epub 2009 Oct 26. Trends Immunol. 2009. PMID: 19864182 Free PMC article. Review.
Cited by
-
Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions.Cell Rep. 2014 Jun 12;7(5):1729-1739. doi: 10.1016/j.celrep.2014.04.052. Epub 2014 May 29. Cell Rep. 2014. PMID: 24882001 Free PMC article.
-
Understanding Viral Transmission Behavior via Protein Intrinsic Disorder Prediction: Coronaviruses.J Pathog. 2012;2012:738590. doi: 10.1155/2012/738590. Epub 2012 Oct 14. J Pathog. 2012. PMID: 23097708 Free PMC article.
-
Nipah shell disorder, modes of infection, and virulence.Microb Pathog. 2020 Apr;141:103976. doi: 10.1016/j.micpath.2020.103976. Epub 2020 Jan 12. Microb Pathog. 2020. PMID: 31940461 Free PMC article.
-
Differences in the number of intrinsically disordered regions between yeast duplicated proteins, and their relationship with functional divergence.PLoS One. 2011;6(9):e24989. doi: 10.1371/journal.pone.0024989. Epub 2011 Sep 15. PLoS One. 2011. PMID: 21949823 Free PMC article.
-
Shell Disorder Models Detect That Omicron Has Harder Shells with Attenuation but Is Not a Descendant of the Wuhan-Hu-1 SARS-CoV-2.Biomolecules. 2022 Apr 25;12(5):631. doi: 10.3390/biom12050631. Biomolecules. 2022. PMID: 35625559 Free PMC article.
References
-
- Brooks GF, Butel JS, Morse SA. Jawetz, Melnick and Adelberg's medical microbiology. 23. New York: Lange/McGraw-Hill; 2004.
-
- Garcia-Sastre A, Whitley RJ. Lessons learned from reconstructing the 1918 influenza pandemic. J Infect Dis. 2006;194:S127–132. - PubMed
-
- Reid AH, Taubenberger JK, Fanning TG. The 1918 Spanish influenza: integrating history and biology. Microbes Infect. 2001;3:81–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

