Epigenetics: connecting environment and genotype to phenotype and disease
- PMID: 19493882
- PMCID: PMC3317936
- DOI: 10.1177/0022034509335868
Epigenetics: connecting environment and genotype to phenotype and disease
Abstract
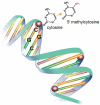
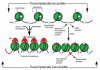
Genetic information is encoded not only by the linear sequence of DNA, but also by epigenetic modifications of chromatin structure that include DNA methylation and covalent modifications of the proteins that bind DNA. These "epigenetic marks" alter the structure of chromatin to influence gene expression. Methylation occurs naturally on cytosine bases at CpG sequences and is involved in controlling the correct expression of genes. DNA methylation is usually associated with triggering histone deacetylation, chromatin condensation, and gene silencing. Differentially methylated cytosines give rise to distinct patterns specific for each tissue type and disease state. Such methylation-variable positions (MVPs) are not uniformly distributed throughout our genome, but are concentrated among genes that regulate transcription, growth, metabolism, differentiation, and oncogenesis. Alterations in MVP methylation status create epigenetic patterns that appear to regulate gene expression profiles during cell differentiation, growth, and development, as well as in cancer. Environmental stressors including toxins, as well as microbial and viral exposures, can change epigenetic patterns and thereby effect changes in gene activation and cell phenotype. Since DNA methylation is often retained following cell division, altered MVP patterns in tissues can accumulate over time and can lead to persistent alterations in steady-state cellular metabolism, responses to stimuli, or the retention of an abnormal phenotype, reflecting a molecular consequence of gene-environment interaction. Hence, DNA epigenetics constitutes the main and previously missing link among genetics, disease, and the environment. The challenge in oral biology will be to understand the mechanisms that modify MVPs in oral tissues and to identify those epigenetic patterns that modify disease pathogenesis or responses to therapy.
Figures
References
-
- Adcock IM, Lee KY. (2006). Abnormal histone acetylase and deacetylase expression and function in lung inflammation. Inflamm Res 55:311-321, erratum in Inflamm Res 55:572, 2006 - PubMed
-
- Ansel KM, Lee DU, Rao A. (2003). An epigenetic view of helper T cell differentiation. Nat Immunol 4:616-623 - PubMed
-
- Barker DJ, Eriksson JG, Forsen T, Osmond C. (2002). Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol 31:1235-1239 - PubMed
-
- Bartolomei MS, Zemel S, Tilghman MS. (1991). Parental imprinting of the mouse H19 gene. Nature 351:153-155 - PubMed
-
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev 16:6-21 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

