Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs
- PMID: 19493973
- PMCID: PMC2719132
- DOI: 10.1104/pp.109.138438
Evidence for variation in the optimal translation initiation complex: plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs
Abstract
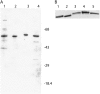
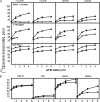
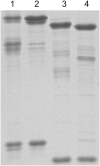
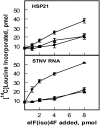
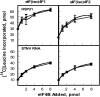
Eukaryotic initiation factor (eIF) 4B is known to interact with multiple initiation factors, mRNA, rRNA, and poly(A) binding protein (PABP). To gain a better understanding of the function of eIF4B, the two isoforms from Arabidopsis (Arabidopsis thaliana) were expressed and analyzed using biophysical and biochemical methods. Plant eIF4B was found by ultracentrifugation and light scattering analysis to most likely be a monomer with an extended structure. An extended structure would facilitate the multiple interactions of eIF4B with mRNA as well as other initiation factors (eIF4A, eIF4G, PABP, and eIF3). Eight mRNAs, barley (Hordeum vulgare) alpha-amylase mRNA, rabbit beta-hemoglobin mRNA, Arabidopsis heat shock protein 21 (HSP21) mRNA, oat (Avena sativa) globulin, wheat (Triticum aestivum) germin, maize (Zea mays) alcohol dehydrogenase, satellite tobacco necrosis virus RNA, and alfalfa mosaic virus (AMV) 4, were used in wheat germ in vitro translation assays to measure their dependence on eIF4B and eIF4F isoforms. The two Arabidopsis eIF4B isoforms, as well as native and recombinant wheat eIF4B, showed similar responses in the translation assay. AMV RNA 4 and Arabidopsis HSP21 showed only a slight dependence on the presence of eIF4B isoforms, whereas rabbit beta-hemoglobin mRNA and wheat germin mRNA showed modest dependence. Barley alpha-amylase, oat globulin, and satellite tobacco necrosis virus RNA displayed the strongest dependence on eIF4B. These results suggest that eIF4B has some effects on mRNA discrimination during initiation of translation. Barley alpha-amylase, oat globulin, and rabbit beta-hemoglobin mRNA showed the highest activity with eIF4F, whereas Arabidopsis HSP21 and AMV RNA 4 used both eIF4F and eIF(iso)4F equally well. These results suggest that differential or optimal translation of mRNAs may require initiation complexes composed of specific isoforms of initiation factor gene products. Thus, individual mRNAs or classes of mRNAs may respond to the relative abundance of a particular initiation factor(s), which in turn may affect the amount of protein translated. It is likely that optimal multifactor initiation complexes exist that allow for optimal translation of mRNAs under a variety of cellular conditions.
Figures
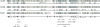

Similar articles
-
Evidence that the 5'-untranslated leader of mRNA affects the requirement for wheat germ initiation factors 4A, 4F, and 4G.J Biol Chem. 1988 Jul 15;263(20):9630-4. J Biol Chem. 1988. PMID: 3260235
-
The 3' mRNA I-shaped structure of maize necrotic streak virus binds to eukaryotic translation factors for eIF4F-mediated translation initiation.J Biol Chem. 2018 Jun 15;293(24):9486-9495. doi: 10.1074/jbc.RA118.003377. Epub 2018 Apr 26. J Biol Chem. 2018. PMID: 29700118 Free PMC article.
-
The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B.J Biol Chem. 2000 Jun 9;275(23):17452-62. doi: 10.1074/jbc.M001186200. J Biol Chem. 2000. PMID: 10747998
-
eIF4F: a retrospective.J Biol Chem. 2015 Oct 2;290(40):24091-9. doi: 10.1074/jbc.R115.675280. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324716 Free PMC article. Review.
-
A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation.Gene. 1998 Aug 17;216(1):1-11. doi: 10.1016/s0378-1119(98)00318-7. Gene. 1998. PMID: 9714706 Review.
Cited by
-
Translation initiation factor AteIF(iso)4E is involved in selective mRNA translation in Arabidopsis thaliana seedlings.PLoS One. 2012;7(2):e31606. doi: 10.1371/journal.pone.0031606. Epub 2012 Feb 20. PLoS One. 2012. PMID: 22363683 Free PMC article.
-
Arabidopsis eIF4E1 protects the translational machinery during TuMV infection and restricts virus accumulation.PLoS Pathog. 2023 Nov 20;19(11):e1011417. doi: 10.1371/journal.ppat.1011417. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37983287 Free PMC article.
-
The role of the poly(A) binding protein in the assembly of the Cap-binding complex during translation initiation in plants.Translation (Austin). 2014 Oct 30;2(2):e959378. doi: 10.4161/2169074X.2014.959378. eCollection 2014 Sep 1. Translation (Austin). 2014. PMID: 26779409 Free PMC article. Review.
-
An Integrated "Multi-Omics" Comparison of Embryo and Endosperm Tissue-Specific Features and Their Impact on Rice Seed Quality.Front Plant Sci. 2017 Nov 22;8:1984. doi: 10.3389/fpls.2017.01984. eCollection 2017. Front Plant Sci. 2017. PMID: 29213276 Free PMC article.
-
Regulation of Germ Cell mRNPs by eIF4E:4EIP Complexes: Multiple Mechanisms, One Goal.Front Cell Dev Biol. 2020 Jul 7;8:562. doi: 10.3389/fcell.2020.00562. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32733883 Free PMC article. Review.
References
-
- Abramson RD, Browning KS, Dever TE, Lawson TG, Thach RE, Ravel JM, Merrick WC (1988. a) Initiation factors that bind mRNA: a comparison of mammalian factors with wheat germ factors. J Biol Chem 263 5462–5467 - PubMed
-
- Abramson RD, Dever TE, Lawson TG, Ray BK, Thach RE, Merrick WC (1987) The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem 262 3826–3832 - PubMed
-
- Abramson RD, Dever TE, Merrick WC (1988. b) Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J Biol Chem 263 6016–6019 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

