The M10 locus of murine gammaherpesvirus 68 contributes to both the lytic and the latent phases of infection
- PMID: 19493995
- PMCID: PMC2715785
- DOI: 10.1128/JVI.00629-09
The M10 locus of murine gammaherpesvirus 68 contributes to both the lytic and the latent phases of infection
Abstract
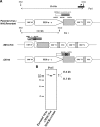
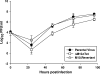
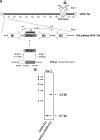
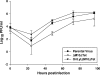
Murine gammaherpesvirus 68 (MHV-68) is closely related to Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus (KSHV) and provides a small-animal model to study the pathogenesis of gammaherpesvirus (gammaHV) infections. According to the colinear organization of the gammaHV genomes, the M10 locus is situated at a position equivalent to the K12 locus of KSHV, which codes for proteins of the kaposin family. The M10 locus of MHV-68 has been predicted to code for three overlapping open reading frames (M10a, M10b, and M10c [M10a-c]) with unknown function. In addition, the M10 locus contains a lytic origin of replication (oriLyt). To elucidate the function of the M10 locus during lytic and latent infections, we investigated, both in vitro and in vivo, the following four recombinant viruses which were generated using MHV-68 cloned as a bacterial artificial chromosome: (i) a mutant virus with a deletion which affects both the coding region for M10a-c and the oriLyt; (ii) a revertant virus in which both the M10a-c coding region and the oriLyt were reverted to those of the wild type; (iii) a virus with an ectopic insertion of the oriLyt, which restores the function of the oriLyt but not the M10a-c coding region; and (iv) a mutant virus with a deletion in the oriLyt only. While the mutants were slightly attenuated with regard to lytic replication in cell culture, they showed severe growth defects in vivo. Both lytic replication and latency amplification were strongly reduced. In contrast, both the revertant virus and the virus with the ectopic oriLyt insertion grew very similarly to the parental wild-type virus both in vitro and in vivo. Thus, we provide genetic evidence that mutation of the oriLyt, and not of putative protein coding sequences within the M10a-c region, is responsible for the observed phenotype. We conclude that the oriLyt in the M10 locus plays an important role during infection of mice with MHV-68.
Figures




Similar articles
-
Establishment of cell lines latently infected with non-oncogenic murine gammaherpesvirus 76.Acta Virol. 2010;54(4):287-91. doi: 10.4149/av_2010_04_287. Acta Virol. 2010. PMID: 21175252
-
Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency.J Virol. 2000 Apr;74(8):3659-67. doi: 10.1128/jvi.74.8.3659-3667.2000. J Virol. 2000. PMID: 10729142 Free PMC article.
-
Murine gammaherpesvirus 68 ORF35 is required for efficient lytic replication and latency.J Gen Virol. 2015 Dec;96(12):3624-3634. doi: 10.1099/jgv.0.000310. J Gen Virol. 2015. PMID: 26459827
-
Gamma herpesviruses: pathogenesis of infection and cell signaling.Folia Microbiol (Praha). 2003;48(3):291-318. doi: 10.1007/BF02931360. Folia Microbiol (Praha). 2003. PMID: 12879740 Review.
-
Murine herpesvirus pathogenesis: a model for the analysis of molecular mechanisms of human gamma herpesvirus infections.Acta Microbiol Immunol Hung. 2005;52(1):41-71. doi: 10.1556/AMicr.52.2005.1.2. Acta Microbiol Immunol Hung. 2005. PMID: 15957234 Review.
Cited by
-
Initiation of lytic DNA replication in Epstein-Barr virus: search for a common family mechanism.Future Virol. 2010 Jan;5(1):65-83. doi: 10.2217/fvl.09.69. Future Virol. 2010. PMID: 22468146 Free PMC article.
-
The small noncoding RNAs (sncRNAs) of murine gammaherpesvirus 68 (MHV-68) are involved in regulating the latent-to-lytic switch in vivo.Sci Rep. 2016 Aug 26;6:32128. doi: 10.1038/srep32128. Sci Rep. 2016. PMID: 27561205 Free PMC article.
-
A Mouse Model to Study the Pathogenesis of γ-herpesviral Infections in Germinal Center B Cells.Cells. 2023 Dec 6;12(24):2780. doi: 10.3390/cells12242780. Cells. 2023. PMID: 38132100 Free PMC article.
-
Nanoparticle exposure reactivates latent herpesvirus and restores a signature of acute infection.Part Fibre Toxicol. 2017 Jan 10;14(1):2. doi: 10.1186/s12989-016-0181-1. Part Fibre Toxicol. 2017. PMID: 28069010 Free PMC article.
-
ORF6 and ORF61 Expressing MVA Vaccines Impair Early but Not Late Latency in Murine Gammaherpesvirus MHV-68 Infection.Front Immunol. 2019 Dec 18;10:2984. doi: 10.3389/fimmu.2019.02984. eCollection 2019. Front Immunol. 2019. PMID: 31921215 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

