The CXCR4-tropic human immunodeficiency virus envelope promotes more-efficient gene delivery to resting CD4+ T cells than the vesicular stomatitis virus glycoprotein G envelope
- PMID: 19493998
- PMCID: PMC2715791
- DOI: 10.1128/JVI.00220-09
The CXCR4-tropic human immunodeficiency virus envelope promotes more-efficient gene delivery to resting CD4+ T cells than the vesicular stomatitis virus glycoprotein G envelope
Abstract
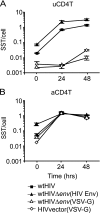
Current gene transfer protocols for resting CD4(+) T cells include an activation step to enhance transduction efficiency. This step is performed because it is thought that resting cells are resistant to transduction by lentiviral-based gene therapy vectors. However, activating resting cells prior to transduction alters their physiology, with foreseeable and unforeseeable negative consequences. Thus, it would be desirable to transduce resting CD4(+) T cells without activation. We recently demonstrated, contrary to the prevailing belief, that wild-type human immunodeficiency virus (HIV) integrates into resting CD4(+) T cells. Based on that finding, we investigated whether a commonly used, vesicular stomatitis virus glycoprotein G (VSV-G)-pseudotyped lentiviral gene therapy vector could also integrate into resting CD4(+) T cells. To investigate this, we inoculated resting CD4(+) T cells with lentiviral particles that were pseudotyped with VSV-G or CXCR4-tropic HIV Env and assayed binding, fusion, reverse transcription, and integration. We found that the VSV-G-pseudotyped lentiviral vector failed to fuse to resting CD4(+) T cells while HIV Env-pseudotyped lentiviral vectors fused, reverse transcribed, and integrated in resting cells. Our findings suggest that HIV Env could be used effectively for the delivery of therapeutic genes to resting CD4(+) T cells and suggest that fusion may be the critical step restricting transduction of resting CD4(+) T cells by lentiviral gene therapy vectors.
Figures





References
-
- Aiuti, A. 2002. Advances in gene therapy for ADA-deficient SCID. Curr. Opin. Mol. Ther. 4515-522. - PubMed
-
- Akkina, R. K., R. M. Walton, M. L. Chen, Q. X. Li, V. Planelles, and I. S. Chen. 1996. High-efficiency gene transfer into CD34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein G. J. Virol. 702581-2585. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

