Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: Evidence that NBs are sites of viral transcription and replication
- PMID: 19494013
- PMCID: PMC2715764
- DOI: 10.1128/JVI.00554-09
Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: Evidence that NBs are sites of viral transcription and replication
Abstract
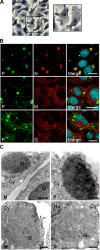
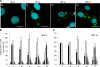
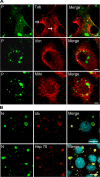
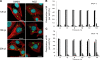
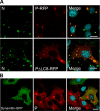
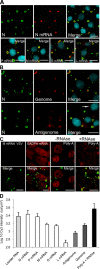
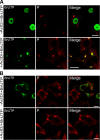
Rabies virus infection induces the formation of cytoplasmic inclusion bodies that resemble Negri bodies found in the cytoplasm of some infected nerve cells. We have studied the morphogenesis and the role of these Negri body-like structures (NBLs) during viral infection. The results indicate that these spherical structures (one or two per cell in the initial stage of infection), composed of the viral N and P proteins, grow during the virus cycle before appearing as smaller structures at late stages of infection. We have shown that the microtubule network is not necessary for the formation of these inclusion bodies but is involved in their dynamics. In contrast, the actin network does not play any detectable role in these processes. These inclusion bodies contain Hsp70 and ubiquitinylated proteins, but they are not misfolded protein aggregates. NBLs, in fact, appear to be functional structures involved in the viral life cycle. Specifically, using in situ fluorescent hybridization techniques, we show that all viral RNAs (genome, antigenome, and every mRNA) are located inside the inclusion bodies. Significantly, short-term RNA labeling in the presence of BrUTP strongly suggests that the NBLs are the sites where viral transcription and replication take place.
Figures


References
-
- Chen, M., and D. Gerlier. 2006. Viral hijacking of cellular ubiquitination pathways as an anti-innate immunity strategy. Viral Immunol. 19349-362. - PubMed
-
- Chenik, M., K. Chebli, Y. Gaudin, and D. Blondel. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75(Pt. 11)2889-2896. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

