Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif
- PMID: 19494014
- PMCID: PMC2715790
- DOI: 10.1128/JVI.00280-09
Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif
Abstract
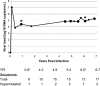
Hypermutation, the introduction of excessive G-to-A substitutions by host proteins in the APOBEC family, can impair replication of the human immunodeficiency virus (HIV). Because hypermutation represents a potential antiviral strategy, it is important to determine whether greater hypermutation is associated with slower disease progression in natural infection. We examined the level of HIV-1 hypermutation among 28 antiretroviral-naive Kenyan women at two times during infection. By examining single-copy gag sequences from proviral DNA, hypermutation was detected in 16 of 28 individuals. Among individuals with any hypermutation, a median of 15% of gag sequences were hypermutated (range, 5 to 43%). However, there was no association between the level of gag hypermutation and the viral load or CD4 count. Thus, we observed no overall relationship between hypermutation and markers of disease progression among individuals with low to moderate levels of hypermutation. In addition, one individual sustained a typical viral load despite having a high level of hypermutation. This individual had 43% of gag sequences hypermutated and harbored a partially defective Vif, which was found to permit hypermutation in a peripheral blood mononuclear cell culture. Overall, our results suggest that a potential antiviral therapy based on hypermutation may need to achieve a substantially higher level of hypermutation than is naturally seen in most individuals to impair virus replication and subsequent disease progression.
Figures

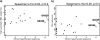


References
-
- Benki, S., R. S. McClelland, S. Emery, J. M. Baeten, B. A. Richardson, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2006. Quantification of genital human immunodeficiency virus type 1 (HIV-1) DNA in specimens from women with low plasma HIV-1 RNA levels typical of HIV-1 nontransmitters. J. Clin. Microbiol. 444357-4362. - PMC - PubMed
-
- Chelico, L., P. Pham, P. Calabrese, and M. F. Goodman. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13392-399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

