Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae
- PMID: 19494041
- PMCID: PMC2719568
- DOI: 10.1091/mbc.e09-02-0111
Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae
Abstract
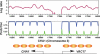
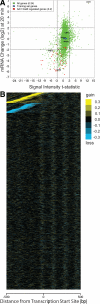
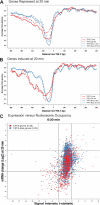
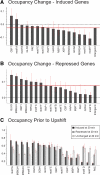
Several well-studied promoters in yeast lose nucleosomes upon transcriptional activation and gain them upon repression, an observation that has prompted the model that transcriptional activation and repression requires nucleosome remodeling of regulated promoters. We have examined global nucleosome positioning before and after glucose-induced transcriptional reprogramming, a condition under which more than half of all yeast genes significantly change expression. The majority of induced and repressed genes exhibit no change in promoter nucleosome arrangement, although promoters that do undergo nucleosome remodeling tend to contain a TATA box. Rather, we found multiple examples where the pre-existing accessibility of putative transcription factor binding sites before glucose addition determined whether the corresponding gene would change expression in response to glucose addition. These results suggest that selection of appropriate transcription factor binding sites may be dictated to a large extent by nucleosome prepositioning but that regulation of expression through these sites is dictated not by nucleosome repositioning but by changes in transcription factor activity.
Figures


References
-
- Agricola E., Verdone L., Xella B., Di Mauro E., Caserta M. Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae. Biochemistry. 2004;43:8878–8884. - PubMed
-
- Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., Schuster S. C., Pugh B. F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Basehoar A. D., Zanton S. J., Pugh B. F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
-
- Beer M. A., Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

